|
probeID |
AGICode |
Annotation |
Log2 signal ratio
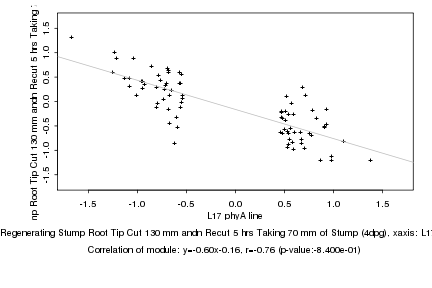
L17 phyA line |
Log2 signal ratio
Regenerating Stump Root Tip Cut 130 mm andn Recut 5 hrs Taking 70 mm of Stump (4dpg) |
| 1 |
264143_at |
AT1G79330
|
AMC6, ATMC5, metacaspase 5, ATMCP2b, metacaspase 2b, MC5, metacaspase 5, MCP2b, metacaspase 2b |
1.377 |
-1.204 |
| 2 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.102 |
-0.806 |
| 3 |
260092_at |
AT1G73280
|
scpl3, serine carboxypeptidase-like 3 |
0.978 |
-1.108 |
| 4 |
253689_at |
AT4G29770
|
unknown |
0.972 |
-1.191 |
| 5 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
0.923 |
-0.153 |
| 6 |
257354_x_at |
AT2G23480
|
unknown |
0.923 |
-0.454 |
| 7 |
254582_at |
AT4G19470
|
[Leucine-rich repeat (LRR) family protein] |
0.908 |
-0.502 |
| 8 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
0.906 |
-0.528 |
| 9 |
267110_at |
AT2G14800
|
unknown |
0.863 |
-1.205 |
| 10 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.822 |
-0.328 |
| 11 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
0.785 |
-0.174 |
| 12 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
0.770 |
-0.684 |
| 13 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.753 |
-0.637 |
| 14 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.707 |
0.141 |
| 15 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
0.696 |
-0.957 |
| 16 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.674 |
0.305 |
| 17 |
250425_at |
AT5G10530
|
LecRK-IX.1, L-type lectin receptor kinase IX.1 |
0.667 |
-0.758 |
| 18 |
258782_at |
AT3G11750
|
FOLB1 |
0.665 |
-0.848 |
| 19 |
259902_at |
AT1G74170
|
AtRLP13, receptor like protein 13, RLP13, receptor like protein 13 |
0.661 |
-0.620 |
| 20 |
257872_at |
AT3G28360
|
ABCB16, ATP-binding cassette B16, PGP16, P-glycoprotein 16 |
0.597 |
-0.617 |
| 21 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.590 |
-0.979 |
| 22 |
244997_at |
ATCG00170
|
RPOC2 |
0.584 |
-0.262 |
| 23 |
262342_at |
AT1G64150
|
[Uncharacterized protein family (UPF0016)] |
0.575 |
-0.825 |
| 24 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.569 |
-0.039 |
| 25 |
250960_at |
AT5G02940
|
unknown |
0.559 |
-0.540 |
| 26 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.546 |
-0.765 |
| 27 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
0.540 |
-0.258 |
| 28 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
0.540 |
-0.873 |
| 29 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.537 |
-0.649 |
| 30 |
262196_at |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
0.530 |
-0.930 |
| 31 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.530 |
-0.609 |
| 32 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.512 |
0.113 |
| 33 |
253211_at |
AT4G34880
|
[Amidase family protein] |
0.505 |
-0.189 |
| 34 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
0.502 |
-0.371 |
| 35 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
0.495 |
-0.558 |
| 36 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.478 |
-0.341 |
| 37 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.476 |
-0.646 |
| 38 |
249127_at |
AT5G43500
|
ARP9, actin-related protein 9, ATARP9, actin-related protein 9 |
0.465 |
-0.197 |
| 39 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
0.462 |
-0.316 |
| 40 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.460 |
-0.205 |
| 41 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.456 |
-0.628 |
| 42 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.680 |
0.644 |
| 43 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-1.255 |
0.605 |
| 44 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.237 |
1.009 |
| 45 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.221 |
0.889 |
| 46 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-1.134 |
0.474 |
| 47 |
264680_at |
AT1G65510
|
unknown |
-1.090 |
0.474 |
| 48 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-1.089 |
0.318 |
| 49 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-1.041 |
0.896 |
| 50 |
246888_at |
AT5G26270
|
unknown |
-1.012 |
0.124 |
| 51 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
-0.968 |
0.415 |
| 52 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.957 |
0.280 |
| 53 |
266282_at |
AT2G29220
|
LecRK-III.1, L-type lectin receptor kinase III.1 |
-0.957 |
0.425 |
| 54 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.938 |
0.361 |
| 55 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.866 |
0.738 |
| 56 |
263673_at |
AT2G04800
|
unknown |
-0.811 |
0.291 |
| 57 |
247868_at |
AT5G57620
|
AtMYB36, myb domain protein 36, MYB36, myb domain protein 36 |
-0.806 |
-0.119 |
| 58 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-0.800 |
-0.029 |
| 59 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.786 |
0.543 |
| 60 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.774 |
0.434 |
| 61 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.738 |
0.054 |
| 62 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.732 |
0.249 |
| 63 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
-0.714 |
0.330 |
| 64 |
252659_at |
AT3G44430
|
unknown |
-0.710 |
0.381 |
| 65 |
248980_at |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
-0.700 |
0.693 |
| 66 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.693 |
0.614 |
| 67 |
262719_at |
AT1G43590
|
unknown |
-0.691 |
-0.147 |
| 68 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.685 |
0.656 |
| 69 |
249010_at |
AT5G44580
|
unknown |
-0.678 |
0.129 |
| 70 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.678 |
-0.447 |
| 71 |
245082_at |
AT2G23270
|
unknown |
-0.655 |
0.230 |
| 72 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.631 |
-0.841 |
| 73 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.606 |
-0.320 |
| 74 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
-0.595 |
-0.514 |
| 75 |
246743_at |
AT5G27750
|
[F-box/FBD-like domains containing protein] |
-0.577 |
0.381 |
| 76 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.577 |
0.600 |
| 77 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.570 |
-0.109 |
| 78 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.563 |
0.375 |
| 79 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.559 |
-0.016 |
| 80 |
257059_at |
AT3G15280
|
unknown |
-0.559 |
0.568 |
| 81 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
-0.549 |
0.143 |
| 82 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
-0.549 |
0.072 |