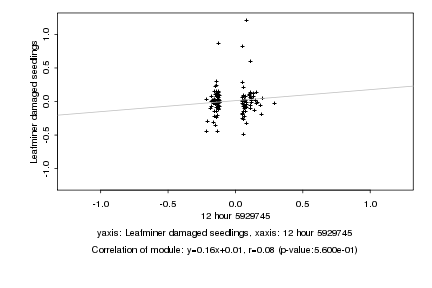
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
12 hour 5929745 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
266767_at |
AT2G46910
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.288 |
-0.023 |
| 2 |
251320_at |
AT3G61530
|
PANB2 |
0.198 |
0.049 |
| 3 |
261301_at |
AT1G48570
|
[zinc finger (Ran-binding) family protein] |
0.193 |
-0.190 |
| 4 |
247561_at |
AT5G61110
|
[zinc ion binding] |
0.179 |
-0.050 |
| 5 |
245581_at |
AT4G14840
|
unknown |
0.157 |
-0.013 |
| 6 |
256615_at |
AT3G22250
|
[UDP-Glycosyltransferase superfamily protein] |
0.154 |
-0.018 |
| 7 |
244921_s_at |
ATMG01000
|
ORF114 |
0.152 |
0.135 |
| 8 |
261072_at |
AT1G07340
|
ATSTP2, sugar transporter 2, STP2, sugar transporter 2 |
0.145 |
0.024 |
| 9 |
267185_at |
AT2G43950
|
ATOEP37, ARABIDOPSIS CHLOROPLAST OUTER ENVELOPE PROTEIN 37, OEP37, chloroplast outer envelope protein 37 |
0.136 |
-0.128 |
| 10 |
264565_at |
AT1G05280
|
unknown |
0.130 |
0.089 |
| 11 |
253868_at |
AT4G27500
|
PPI1, proton pump interactor 1 |
0.129 |
0.128 |
| 12 |
266928_at |
AT2G45790
|
ATPMM, PHOSPHOMANNOMUTASE, PMM, phosphomannomutase |
0.119 |
-0.012 |
| 13 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
0.117 |
0.057 |
| 14 |
267568_at |
AT2G30780
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.116 |
0.029 |
| 15 |
263949_at |
AT2G36060
|
MMZ3, MMS ZWEI homologue 3, UEV1C, UBIQUITIN E2 VARIANT 1C |
0.112 |
0.049 |
| 16 |
248631_at |
AT5G49000
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.111 |
0.061 |
| 17 |
262744_at |
AT1G28680
|
[HXXXD-type acyl-transferase family protein] |
0.110 |
0.131 |
| 18 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.108 |
0.598 |
| 19 |
254403_at |
AT4G21323
|
[Subtilase family protein] |
0.106 |
-0.047 |
| 20 |
251249_at |
AT3G62160
|
[HXXXD-type acyl-transferase family protein] |
0.106 |
-0.096 |
| 21 |
248259_at |
AT5G53330
|
[Ubiquitin-associated/translation elongation factor EF1B protein] |
0.106 |
0.143 |
| 22 |
263687_at |
AT1G26940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.100 |
0.101 |
| 23 |
264522_at |
AT1G10050
|
[glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein] |
0.099 |
0.118 |
| 24 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.081 |
1.222 |
| 25 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
0.081 |
-0.316 |
| 26 |
267273_at |
AT2G02520
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
0.081 |
-0.002 |
| 27 |
253473_at |
AT4G32250
|
[Protein kinase superfamily protein] |
0.079 |
-0.041 |
| 28 |
248023_at |
AT5G56450
|
PM-ANT |
0.076 |
-0.081 |
| 29 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
0.072 |
-0.147 |
| 30 |
255299_at |
AT4G04880
|
[adenosine/AMP deaminase family protein] |
0.072 |
-0.080 |
| 31 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.071 |
0.082 |
| 32 |
257574_at |
AT3G20710
|
[F-box family protein] |
0.070 |
0.029 |
| 33 |
253272_at |
AT4G34190
|
SEP1, stress enhanced protein 1 |
0.069 |
-0.068 |
| 34 |
267405_at |
AT2G33740
|
CUTA |
0.066 |
0.006 |
| 35 |
246265_at |
AT1G31860
|
AT-IE, HISN2, HISTIDINE BIOSYNTHESIS 2 |
0.065 |
-0.215 |
| 36 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
0.063 |
-0.091 |
| 37 |
247969_at |
AT5G56700
|
[FBD / Leucine Rich Repeat domains containing protein] |
0.062 |
0.038 |
| 38 |
250291_at |
AT5G13280
|
AK, ASPARTATE KINASE, AK-LYS1, aspartate kinase 1, AK1, ASPARTATE KINASE 1 |
0.060 |
-0.074 |
| 39 |
253313_at |
AT4G33870
|
[Peroxidase superfamily protein] |
0.060 |
-0.057 |
| 40 |
251829_at |
AT3G55020
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.059 |
-0.146 |
| 41 |
251327_at |
AT3G61540
|
[alpha/beta-Hydrolases superfamily protein] |
0.059 |
0.095 |
| 42 |
247198_at |
AT5G65290
|
[LMBR1-like membrane protein] |
0.057 |
0.080 |
| 43 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.055 |
-0.480 |
| 44 |
246995_at |
AT5G67470
|
ATFH6, ARABIDOPSIS FORMIN HOMOLOG 6, FH6, formin homolog 6 |
0.054 |
-0.261 |
| 45 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.054 |
-0.067 |
| 46 |
252050_at |
AT3G52550
|
unknown |
0.053 |
-0.040 |
| 47 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.053 |
0.219 |
| 48 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.052 |
0.828 |
| 49 |
250322_at |
AT5G12870
|
ATMYB46, MYB DOMAIN PROTEIN 46, MYB46, myb domain protein 46 |
0.051 |
-0.017 |
| 50 |
254754_at |
AT4G13210
|
[Pectin lyase-like superfamily protein] |
0.051 |
0.002 |
| 51 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.051 |
0.294 |
| 52 |
254918_at |
AT4G11260
|
ATSGT1B, EDM1, ENHANCED DOWNY MILDEW 1, ETA3, ENHANCER OF TIR1-1 AUXIN RESISTANCE 3, RPR1, SGT1B |
0.050 |
0.073 |
| 53 |
257706_at |
AT3G12685
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.048 |
-0.253 |
| 54 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
0.047 |
-0.179 |
| 55 |
256728_at |
AT3G25660
|
[Amidase family protein] |
0.047 |
-0.168 |
| 56 |
258140_at |
AT3G24503
|
ALDH1A, aldehyde dehydrogenase 1A, ALDH2C4, aldehyde dehydrogenase 2C4, REF1, REDUCED EPIDERMAL FLUORESCENCE1 |
0.046 |
0.023 |
| 57 |
249837_at |
AT5G23480
|
[SWIB/MDM2 domain] |
-0.217 |
-0.446 |
| 58 |
248357_at |
AT5G52380
|
[VASCULAR-RELATED NAC-DOMAIN 6] |
-0.216 |
0.043 |
| 59 |
262377_at |
AT1G73110
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.211 |
-0.291 |
| 60 |
256926_at |
AT3G22540
|
unknown |
-0.192 |
-0.100 |
| 61 |
245779_at |
AT1G73510
|
unknown |
-0.185 |
0.082 |
| 62 |
249503_at |
AT5G39310
|
ATEXP24, ATEXPA24, expansin A24, ATHEXP ALPHA 1.19, EXP24, EXPANSIN 24, EXPA24, expansin A24 |
-0.181 |
0.014 |
| 63 |
261759_at |
AT1G15590
|
unknown |
-0.178 |
-0.063 |
| 64 |
264917_at |
AT1G60500
|
DRP4C, Dynamin related protein 4C |
-0.176 |
0.018 |
| 65 |
264762_at |
AT1G61460
|
[S-locus protein kinase, putative] |
-0.166 |
-0.019 |
| 66 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.165 |
-0.311 |
| 67 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.163 |
0.023 |
| 68 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.161 |
-0.137 |
| 69 |
264981_at |
AT1G27160
|
[valyl-tRNA synthetase / valine--tRNA ligase-related] |
-0.161 |
-0.010 |
| 70 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.160 |
0.032 |
| 71 |
263000_at |
AT1G54350
|
ABCD2, ATP-binding cassette D2 |
-0.158 |
-0.221 |
| 72 |
245310_at |
AT4G13950
|
RALFL31, ralf-like 31 |
-0.156 |
0.150 |
| 73 |
263541_at |
AT2G24860
|
[DnaJ/Hsp40 cysteine-rich domain superfamily protein] |
-0.155 |
0.030 |
| 74 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.155 |
-0.348 |
| 75 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
-0.153 |
-0.032 |
| 76 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.153 |
0.237 |
| 77 |
262845_at |
AT1G14740
|
TTA1, TITANIA 1 |
-0.151 |
0.106 |
| 78 |
247375_at |
AT5G63135
|
unknown |
-0.147 |
0.124 |
| 79 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
-0.146 |
-0.147 |
| 80 |
264094_at |
AT1G79200
|
SCI1, stigma/style cell-cycle inhibitor 1 |
-0.144 |
0.077 |
| 81 |
256049_at |
AT1G07010
|
AtSLP1, SLP1, Shewenella-like protein phosphatase 1 |
-0.144 |
-0.234 |
| 82 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
-0.143 |
0.160 |
| 83 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
-0.143 |
0.310 |
| 84 |
245873_at |
AT1G26260
|
CIB5, cryptochrome-interacting basic-helix-loop-helix 5 |
-0.142 |
-0.017 |
| 85 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
-0.141 |
0.245 |
| 86 |
266614_at |
AT2G14910
|
unknown |
-0.141 |
-0.077 |
| 87 |
257205_at |
AT3G16520
|
UGT88A1, UDP-glucosyl transferase 88A1 |
-0.140 |
-0.086 |
| 88 |
261171_at |
AT1G04880
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.138 |
0.042 |
| 89 |
254874_at |
AT4G11570
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.137 |
-0.017 |
| 90 |
260645_at |
AT1G53300
|
TTL1, tetratricopeptide-repeat thioredoxin-like 1 |
-0.136 |
-0.196 |
| 91 |
262441_at |
AT1G47720
|
OSB1, Organellar Single-stranded |
-0.135 |
-0.012 |
| 92 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
-0.134 |
-0.443 |
| 93 |
258656_at |
AT3G09900
|
ATRAB8E, ATRABE1E, RAB GTPase homolog E1E, RABE1e, RAB GTPase homolog E1E |
-0.134 |
0.085 |
| 94 |
262710_at |
AT1G16210
|
unknown |
-0.133 |
0.067 |
| 95 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.133 |
-0.097 |
| 96 |
257734_at |
AT3G18370
|
ATSYTF, NTMC2T3, NTMC2TYPE3, SYTF |
-0.132 |
0.110 |
| 97 |
258047_at |
AT3G21240
|
4CL2, 4-coumarate:CoA ligase 2, AT4CL2 |
-0.131 |
0.161 |
| 98 |
248718_at |
AT5G47770
|
FPS1, farnesyl diphosphate synthase 1 |
-0.130 |
-0.105 |
| 99 |
257711_at |
AT3G27430
|
PBB1 |
-0.129 |
0.039 |
| 100 |
245652_at |
AT4G13960
|
[F-box/RNI-like superfamily protein] |
-0.129 |
-0.029 |
| 101 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
-0.128 |
0.866 |
| 102 |
266845_at |
AT2G26110
|
unknown |
-0.128 |
-0.109 |
| 103 |
248294_at |
AT5G53060
|
RCF3, regulator of CBF gene expression 3, SHI1, SHINY 1 |
-0.127 |
0.137 |
| 104 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.126 |
-0.048 |
| 105 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.126 |
0.007 |
| 106 |
263907_at |
AT2G36270
|
ABI5, ABA INSENSITIVE 5, AtABI5, GIA1, GROWTH-INSENSITIVITY TO ABA 1 |
-0.126 |
0.033 |
| 107 |
256900_at |
AT3G24670
|
[Pectin lyase-like superfamily protein] |
-0.125 |
-0.073 |
| 108 |
253275_at |
AT4G34215
|
unknown |
-0.124 |
-0.008 |
| 109 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.124 |
0.017 |
| 110 |
262463_at |
AT1G50310
|
ATSTP9, SUGAR TRANSPORTER 9, STP9, sugar transporter 9 |
-0.124 |
-0.011 |
| 111 |
246915_at |
AT5G25880
|
ATNADP-ME3, Arabidopsis thaliana NADP-malic enzyme 3, NADP-ME3, NADP-malic enzyme 3 |
-0.123 |
0.092 |