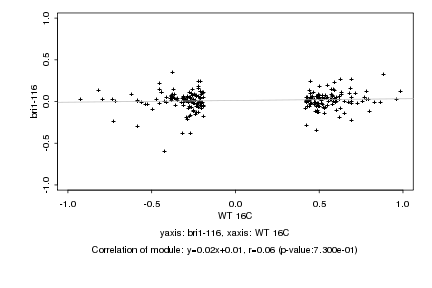
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT 16C |
Log2 signal ratio
bri1-116 |
| 1 |
247424_at |
AT5G62850
|
AtSWEET5, AtVEX1, VEGETATIVE CELL EXPRESSED1, SWEET5 |
0.983 |
0.123 |
| 2 |
260628_at |
AT1G62320
|
[ERD (early-responsive to dehydration stress) family protein] |
0.959 |
0.029 |
| 3 |
253875_at |
AT4G27520
|
AtENODL2, ENODL2, early nodulin-like protein 2 |
0.879 |
0.324 |
| 4 |
250678_at |
AT5G06540
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.860 |
-0.009 |
| 5 |
262956_at |
AT1G54270
|
EIF4A-2, eif4a-2 |
0.830 |
-0.004 |
| 6 |
253892_at |
AT4G27620
|
unknown |
0.797 |
-0.108 |
| 7 |
247564_at |
AT5G61140
|
[U5 small nuclear ribonucleoprotein helicase] |
0.794 |
0.028 |
| 8 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
0.778 |
0.130 |
| 9 |
248957_at |
AT5G45620
|
[Proteasome component (PCI) domain protein] |
0.778 |
0.035 |
| 10 |
255063_at |
AT4G08970
|
[Mutator-like transposase family, has a 8.1e-28 P-value blast match to Q9SL18 /349-510 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.765 |
0.058 |
| 11 |
245926_at |
AT5G24740
|
SHBY, SHRUBBY |
0.755 |
0.012 |
| 12 |
247807_at |
AT5G58360
|
ATOFP3, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 3, OFP3, ovate family protein 3 |
0.725 |
-0.023 |
| 13 |
265613_at |
AT2G25560
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.715 |
0.105 |
| 14 |
258440_at |
AT3G17250
|
[Protein phosphatase 2C family protein] |
0.690 |
-0.020 |
| 15 |
264489_at |
AT1G27370
|
SPL10, squamosa promoter binding protein-like 10 |
0.690 |
0.054 |
| 16 |
254845_at |
AT4G11820
|
FKP1, FLAKY POLLEN 1, HMGS, HYDROXYMETHYLGLUTARYL-COA SYNTHASE, MVA1 |
0.689 |
-0.222 |
| 17 |
255162_at |
AT4G07840
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:BAA22288 pol polyprotein (Ty1_Copia-element) (Oryza australiensis)GB:BAA22288 polyprotein (Ty1_Copia-element) (Oryza australiensis)gi|2443320|dbj|BAA22288.1| polyprotein (RIRE1) (Oryza australiensis) (Ty1_Copia-element)] |
0.688 |
0.002 |
| 18 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
0.688 |
0.273 |
| 19 |
260989_at |
AT1G53450
|
unknown |
0.685 |
0.163 |
| 20 |
247845_at |
AT5G58090
|
[O-Glycosyl hydrolases family 17 protein] |
0.680 |
0.105 |
| 21 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.674 |
-0.007 |
| 22 |
265071_at |
AT1G55520
|
ATTBP2, A. THALIANA TATA BINDING PROTEIN 2, TBP2, TATA binding protein 2 |
0.650 |
0.012 |
| 23 |
267207_at |
AT2G30840
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.647 |
-0.139 |
| 24 |
255978_at |
AT1G34010
|
unknown |
0.632 |
0.113 |
| 25 |
267183_at |
AT2G44020
|
[Mitochondrial transcription termination factor family protein] |
0.631 |
0.089 |
| 26 |
260632_at |
AT1G62360
|
BUM, BUMBERSHOOT, BUM1, BUMBERSHOOT 1, SHL, SHOOTLESS, STM, SHOOT MERISTEMLESS, WAM, WALDMEISTER, WAM1, WALDMEISTER 1 |
0.623 |
0.264 |
| 27 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
0.622 |
-0.074 |
| 28 |
250335_at |
AT5G11650
|
[alpha/beta-Hydrolases superfamily protein] |
0.621 |
-0.181 |
| 29 |
252779_at |
AT3G42990
|
unknown |
0.620 |
0.025 |
| 30 |
260526_at |
AT2G47410
|
[WD40/YVTN repeat-like-containing domain] |
0.619 |
0.067 |
| 31 |
246905_at |
AT5G25570
|
unknown |
0.601 |
0.004 |
| 32 |
254618_at |
AT4G18780
|
ATCESA8, CELLULOSE SYNTHASE 8, CESA8, CELLULOSE SYNTHASE 8, IRX1, IRREGULAR XYLEM 1, LEW2, LEAF WILTING 2 |
0.601 |
-0.099 |
| 33 |
254958_at |
AT4G11010
|
NDPK3, nucleoside diphosphate kinase 3 |
0.600 |
0.030 |
| 34 |
251822_at |
AT3G55060
|
unknown |
0.600 |
0.060 |
| 35 |
259124_at |
AT3G02310
|
AGL4, AGAMOUS-like 4, SEP2, SEPALLATA 2 |
0.597 |
0.006 |
| 36 |
264894_at |
AT1G23040
|
[hydroxyproline-rich glycoprotein family protein] |
0.596 |
-0.010 |
| 37 |
249703_at |
AT5G35560
|
[DENN (AEX-3) domain-containing protein] |
0.594 |
0.011 |
| 38 |
250291_at |
AT5G13280
|
AK, ASPARTATE KINASE, AK-LYS1, aspartate kinase 1, AK1, ASPARTATE KINASE 1 |
0.590 |
0.137 |
| 39 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.588 |
0.238 |
| 40 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.586 |
0.037 |
| 41 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.584 |
0.011 |
| 42 |
260428_at |
AT1G72340
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
0.581 |
0.152 |
| 43 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
0.578 |
0.079 |
| 44 |
245311_at |
AT4G14320
|
[Zinc-binding ribosomal protein family protein] |
0.575 |
0.061 |
| 45 |
247550_at |
AT5G61370
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.575 |
-0.033 |
| 46 |
263286_at |
AT2G36160
|
[Ribosomal protein S11 family protein] |
0.572 |
0.086 |
| 47 |
266903_at |
AT2G34570
|
MEE21, maternal effect embryo arrest 21 |
0.571 |
0.027 |
| 48 |
259860_at |
AT1G80640
|
[Protein kinase superfamily protein] |
0.568 |
0.152 |
| 49 |
253718_at |
AT4G29450
|
[Leucine-rich repeat protein kinase family protein] |
0.566 |
0.030 |
| 50 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.553 |
0.011 |
| 51 |
258690_at |
AT3G07960
|
PIP5K6, phosphatidylinositol-4-phosphate 5-kinase 6 |
0.549 |
-0.046 |
| 52 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
0.549 |
0.196 |
| 53 |
252815_at |
AT3G42540
|
unknown |
0.549 |
0.054 |
| 54 |
255375_at |
AT4G03780
|
[gypsy-like retrotransposon family (Athila), has a 1.9e-59 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.539 |
0.048 |
| 55 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
0.534 |
0.080 |
| 56 |
257269_at |
AT3G17440
|
ATNPSN13, NPSN13, novel plant snare 13 |
0.532 |
-0.071 |
| 57 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.530 |
-0.079 |
| 58 |
264484_at |
AT1G77260
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.528 |
-0.132 |
| 59 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.527 |
0.038 |
| 60 |
262786_at |
AT1G10740
|
[alpha/beta-Hydrolases superfamily protein] |
0.523 |
-0.065 |
| 61 |
245420_at |
AT4G17410
|
[DWNN domain, a CCHC-type zinc finger] |
0.516 |
-0.033 |
| 62 |
248730_at |
AT5G48050
|
unknown |
0.515 |
0.082 |
| 63 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.514 |
0.052 |
| 64 |
262478_at |
AT1G11170
|
unknown |
0.511 |
-0.023 |
| 65 |
258460_at |
AT3G17330
|
ECT6, evolutionarily conserved C-terminal region 6 |
0.511 |
-0.015 |
| 66 |
256038_at |
AT1G19170
|
[Pectin lyase-like superfamily protein] |
0.510 |
0.015 |
| 67 |
267514_at |
AT2G45630
|
[D-isomer specific 2-hydroxyacid dehydrogenase family protein] |
0.505 |
0.032 |
| 68 |
263743_at |
AT2G21390
|
[Coatomer, alpha subunit] |
0.503 |
0.036 |
| 69 |
245580_at |
AT4G14820
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.501 |
0.056 |
| 70 |
250637_at |
AT5G07530
|
ATGRP-7, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 7, ATGRP17, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 17, GRP17, glycine rich protein 17 |
0.497 |
0.188 |
| 71 |
262361_at |
AT1G73150
|
GTE3, global transcription factor group E3 |
0.496 |
-0.053 |
| 72 |
247106_at |
AT5G66240
|
[Transducin/WD40 repeat-like superfamily protein] |
0.496 |
0.073 |
| 73 |
246450_at |
AT5G16820
|
ATHSF3, ARABIDOPSIS HEAT SHOCK FACTOR 3, ATHSFA1B, ARABIDOPSIS THALIANA CLASS A HEAT SHOCK FACTOR 1B, HSF3, heat shock factor 3, HSFA1B, CLASS A HEAT SHOCK FACTOR 1B |
0.495 |
0.040 |
| 74 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
0.494 |
-0.131 |
| 75 |
267407_at |
AT2G34880
|
JMJ15, Jumonji domain-containing protein 15, MEE27, maternal effect embryo arrest 27, PKDM7C |
0.494 |
0.040 |
| 76 |
259118_at |
AT3G01310
|
[Phosphoglycerate mutase-like family protein] |
0.492 |
0.093 |
| 77 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
0.492 |
-0.051 |
| 78 |
256971_at |
AT3G21100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.491 |
-0.013 |
| 79 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
0.489 |
-0.125 |
| 80 |
254170_at |
AT4G24430
|
[Rhamnogalacturonate lyase family protein] |
0.486 |
0.070 |
| 81 |
257422_at |
AT1G11940
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.485 |
-0.036 |
| 82 |
259673_at |
AT1G77800
|
[PHD finger family protein] |
0.485 |
-0.106 |
| 83 |
262514_at |
AT1G34190
|
anac017, NAC domain containing protein 17, NAC017, NAC domain containing protein 17 |
0.484 |
-0.021 |
| 84 |
262955_at |
AT1G54520
|
unknown |
0.483 |
0.072 |
| 85 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.483 |
-0.339 |
| 86 |
248056_at |
AT5G55500
|
ATXYLT, ARABIDOPSIS THALIANA BETA-1,2-XYLOSYLTRANSFERASE, XYLT, beta-1,2-xylosyltransferase |
0.483 |
-0.057 |
| 87 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
0.477 |
-0.110 |
| 88 |
249314_at |
AT5G41180
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.477 |
-0.011 |
| 89 |
252083_at |
AT3G51960
|
ATBZIP24, basic leucine zipper 24, BZIP24, basic leucine zipper 24 |
0.471 |
-0.001 |
| 90 |
267269_at |
AT2G02560
|
ATCAND1, CULLIN-ASSOCIATED AND NEDDYLATION DISSOCIATED 1, CAND1, cullin-associated and neddylation dissociated, ETA2, HVE, HEMIVENATA, TIP120 |
0.470 |
-0.026 |
| 91 |
252398_at |
AT3G47990
|
SIS3, SUGAR-INSENSITIVE 3 |
0.469 |
0.026 |
| 92 |
248593_at |
AT5G49180
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.463 |
0.110 |
| 93 |
258115_at |
AT3G14670
|
unknown |
0.457 |
0.070 |
| 94 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.454 |
-0.022 |
| 95 |
265684_at |
AT2G24410
|
unknown |
0.453 |
0.057 |
| 96 |
252264_at |
AT3G49590
|
ATG13, autophagy-related 13 |
0.449 |
0.045 |
| 97 |
258128_at |
AT3G24590
|
PLSP1, plastidic type i signal peptidase 1 |
0.448 |
0.098 |
| 98 |
252108_at |
AT3G51530
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.446 |
0.057 |
| 99 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
0.446 |
0.052 |
| 100 |
245720_at |
AT5G04210
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
0.445 |
-0.028 |
| 101 |
257777_x_at |
AT3G29210
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
0.445 |
-0.012 |
| 102 |
249175_at |
AT5G42970
|
ATS4, COP14, CONSTITUTIVE PHOTOMORPHOGENIC 14, COP8, CONSTITUTIVE PHOTOMORPHOGENIC 8, CSN4, COP9 SIGNALOSOME SUBUNIT 4, EMB134, EMBRYO DEFECTIVE 134, FUS4, FUSCA 4, FUS8, FUSCA 8 |
0.445 |
0.031 |
| 103 |
258766_at |
AT3G10700
|
GalAK, galacturonic acid kinase |
0.444 |
0.252 |
| 104 |
246623_at |
AT1G48920
|
ATNUC-L1, nucleolin like 1, NUC-L1, nucleolin like 1, PARL1, PARALLEL 1 |
0.443 |
0.047 |
| 105 |
262754_at |
AT1G16350
|
[Aldolase-type TIM barrel family protein] |
0.440 |
0.138 |
| 106 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.440 |
-0.055 |
| 107 |
261354_at |
AT1G79690
|
atnudt3, nudix hydrolase homolog 3, NUDT3, nudix hydrolase homolog 3 |
0.437 |
0.017 |
| 108 |
257265_at |
AT3G14980
|
IDM1, increased DNA methylation 1, ROS4, REPRESSOR OF SILENCING 4 |
0.435 |
0.012 |
| 109 |
245153_at |
AT5G12450
|
[FBD-like domain family protein] |
0.432 |
0.009 |
| 110 |
253952_at |
AT4G26840
|
ATSUMO1, ARABIDOPSIS THALIANA SMALL UBIQUITIN-LIKE MODIFIER 1, SUM1, SMALL UBIQUITIN-LIKE MODIFIER 1, SUMO 1, SMALL UBIQUITIN-LIKE MODIFIER 1, SUMO1, small ubiquitin-like modifier 1 |
0.429 |
0.051 |
| 111 |
249284_at |
AT5G41810
|
unknown |
0.427 |
-0.051 |
| 112 |
263835_at |
AT2G40290
|
[Eukaryotic translation initiation factor 2 subunit 1] |
0.426 |
0.018 |
| 113 |
259233_at |
AT3G11380
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.425 |
0.031 |
| 114 |
266816_at |
AT2G44970
|
[alpha/beta-Hydrolases superfamily protein] |
0.424 |
0.010 |
| 115 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
0.424 |
-0.009 |
| 116 |
246256_at |
AT4G36770
|
[UDP-Glycosyltransferase superfamily protein] |
0.422 |
0.016 |
| 117 |
259319_at |
AT3G01090
|
AKIN10, SNF1 kinase homolog 10, KIN10, KIN10, SNF1 kinase homolog 10, SNRK1.1, SNF1-RELATED PROTEIN KINASE 1.1 |
0.422 |
0.052 |
| 118 |
251444_at |
AT3G59990
|
MAP2B, methionine aminopeptidase 2B |
0.420 |
0.005 |
| 119 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.419 |
-0.277 |
| 120 |
263900_at |
AT2G36290
|
[alpha/beta-Hydrolases superfamily protein] |
0.417 |
-0.076 |
| 121 |
248432_at |
AT5G51390
|
unknown |
-0.928 |
0.033 |
| 122 |
248538_at |
AT5G50110
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.822 |
0.141 |
| 123 |
250334_at |
AT5G11770
|
[NADH-ubiquinone oxidoreductase 20 kDa subunit, mitochondrial] |
-0.795 |
0.033 |
| 124 |
249166_at |
AT5G42840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.739 |
0.025 |
| 125 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.732 |
-0.237 |
| 126 |
263362_at |
AT2G03880
|
REME1, required for efficiency of mitochondrial editing 1 |
-0.720 |
0.005 |
| 127 |
257665_at |
AT3G20430
|
unknown |
-0.623 |
0.085 |
| 128 |
255077_at |
AT4G09150
|
[T-complex protein 11] |
-0.589 |
0.009 |
| 129 |
267410_at |
AT2G34920
|
EDA18, embryo sac development arrest 18 |
-0.588 |
-0.289 |
| 130 |
259436_at |
AT1G01500
|
[Erythronate-4-phosphate dehydrogenase family protein] |
-0.587 |
0.019 |
| 131 |
257670_at |
AT3G20340
|
unknown |
-0.561 |
-0.010 |
| 132 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.542 |
-0.030 |
| 133 |
265461_at |
AT2G46500
|
ATPI4K GAMMA 4, phosphoinositide 4-kinase gamma 4, PI4K GAMMA 4, phosphoinositide 4-kinase gamma 4, UBDK GAMMA 4, UBIQUITIN-LIKE DOMAIN KINASE GAMMA 4 |
-0.530 |
-0.027 |
| 134 |
260646_at |
AT1G53340
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.498 |
-0.084 |
| 135 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.474 |
0.026 |
| 136 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.459 |
0.147 |
| 137 |
265748_at |
AT2G10620
|
[gypsy-like retrotransposon family (Athila), has a 5.5e-59 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.456 |
-0.012 |
| 138 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
-0.455 |
0.226 |
| 139 |
244901_at |
ATMG00640
|
ORF25 |
-0.444 |
0.118 |
| 140 |
247377_at |
AT5G63180
|
[Pectin lyase-like superfamily protein] |
-0.442 |
0.114 |
| 141 |
256926_at |
AT3G22540
|
unknown |
-0.426 |
-0.598 |
| 142 |
264521_at |
AT1G10020
|
unknown |
-0.424 |
0.002 |
| 143 |
259959_at |
AT1G53720
|
ATCYP59, CYCLOPHILIN 59, CYP59, cyclophilin 59 |
-0.415 |
0.050 |
| 144 |
244906_at |
ATMG00690
|
ORF240A |
-0.415 |
-0.010 |
| 145 |
265745_at |
AT2G06640
|
[pseudogene] |
-0.389 |
0.048 |
| 146 |
252142_at |
AT3G51120
|
[DNA binding] |
-0.386 |
0.083 |
| 147 |
257682_at |
AT3G13240
|
unknown |
-0.384 |
0.016 |
| 148 |
252071_at |
AT3G51690
|
[PIF1 helicase] |
-0.382 |
0.061 |
| 149 |
266904_at |
AT2G34590
|
[Transketolase family protein] |
-0.381 |
0.086 |
| 150 |
262214_at |
AT1G74820
|
[RmlC-like cupins superfamily protein] |
-0.376 |
0.038 |
| 151 |
261496_at |
AT1G28360
|
ATERF12, ERF DOMAIN PROTEIN 12, ERF12, ERF domain protein 12 |
-0.376 |
0.354 |
| 152 |
250570_at |
AT5G08170
|
ATAIH, AGMATINE IMINOHYDROLASE, EMB1873, EMBRYO DEFECTIVE 1873 |
-0.375 |
0.057 |
| 153 |
253986_at |
AT4G26210
|
[Mitochondrial ATP synthase subunit G protein] |
-0.372 |
0.145 |
| 154 |
261683_at |
AT1G47300
|
[F-box family protein] |
-0.369 |
0.090 |
| 155 |
246369_at |
AT1G51910
|
[Leucine-rich repeat protein kinase family protein] |
-0.368 |
0.041 |
| 156 |
244902_at |
ATMG00650
|
NAD4L, NADH dehydrogenase subunit 4L |
-0.359 |
-0.042 |
| 157 |
259096_at |
AT3G04840
|
[Ribosomal protein S3Ae] |
-0.353 |
0.036 |
| 158 |
265558_at |
AT2G05550
|
[non-LTR retrotransposon family (LINE), has a 6.6e-37 P-value blast match to GB:NP_038605 L1 repeat, Tf subfamily, member 30 (LINE-element) (Mus musculus)] |
-0.350 |
0.040 |
| 159 |
250439_at |
AT5G10450
|
14-3-3lambda, 14-3-3 PROTEIN G-BOX FACTOR14 LAMBDA, AFT1, GRF6, G-box regulating factor 6 |
-0.349 |
0.014 |
| 160 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
-0.325 |
-0.008 |
| 161 |
265327_at |
AT2G18210
|
unknown |
-0.322 |
-0.381 |
| 162 |
249990_at |
AT5G18540
|
unknown |
-0.317 |
0.041 |
| 163 |
266583_at |
AT2G46220
|
[Uncharacterized conserved protein (DUF2358)] |
-0.315 |
0.060 |
| 164 |
256239_at |
AT3G12470
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.314 |
-0.008 |
| 165 |
248591_at |
AT5G49650
|
XK-2, xylulose kinase-2, XK2, XYLULOSE KINASE 2 |
-0.314 |
0.068 |
| 166 |
255881_at |
AT1G67070
|
DIN9, DARK INDUCIBLE 9, PMI2, PHOSPHOMANNOSE ISOMERASE 2 |
-0.313 |
-0.042 |
| 167 |
260131_at |
AT1G66310
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.308 |
0.048 |
| 168 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
-0.307 |
0.033 |
| 169 |
251801_at |
AT3G55440
|
ATCTIMC, CYTOSOLIC TRIOSE PHOSPHATE ISOMERASE, CYTOTPI, CYTOSOLIC ISOFORM TRIOSE PHOSPHATE ISOMERASE, TPI, triosephosphate isomerase |
-0.305 |
0.020 |
| 170 |
264425_at |
AT1G61750
|
[Receptor-like protein kinase-related family protein] |
-0.298 |
0.001 |
| 171 |
260210_at |
AT1G74420
|
ATFUT3, FUT3, fucosyltransferase 3 |
-0.295 |
-0.190 |
| 172 |
254071_at |
AT4G25510
|
unknown |
-0.293 |
0.013 |
| 173 |
267146_at |
AT2G38160
|
unknown |
-0.291 |
0.017 |
| 174 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
-0.291 |
0.004 |
| 175 |
257894_at |
AT3G17100
|
AIF3, ATBS1 Interacting Factor 3 |
-0.289 |
0.060 |
| 176 |
259309_at |
AT3G05050
|
[Protein kinase superfamily protein] |
-0.288 |
-0.215 |
| 177 |
249549_at |
AT5G38180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.287 |
0.063 |
| 178 |
256696_at |
AT3G20650
|
[mRNA capping enzyme family protein] |
-0.284 |
-0.001 |
| 179 |
259415_at |
AT1G02330
|
unknown |
-0.282 |
-0.077 |
| 180 |
267268_at |
AT2G02570
|
[nucleic acid binding] |
-0.279 |
-0.049 |
| 181 |
263571_at |
AT2G17050
|
[disease resistance protein (TIR-NBS-LRR class), putative] |
-0.278 |
0.053 |
| 182 |
250301_at |
AT5G11970
|
unknown |
-0.277 |
-0.178 |
| 183 |
247890_at |
AT5G57940
|
ATCNGC5, cyclic nucleotide gated channel 5, CNGC5, cyclic nucleotide gated channel 5 |
-0.277 |
0.110 |
| 184 |
260894_at |
AT1G29220
|
[transcriptional regulator family protein] |
-0.274 |
0.016 |
| 185 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.273 |
-0.373 |
| 186 |
259368_at |
AT1G69100
|
[Eukaryotic aspartyl protease family protein] |
-0.270 |
0.052 |
| 187 |
257621_at |
AT3G20410
|
CPK9, calmodulin-domain protein kinase 9 |
-0.269 |
-0.157 |
| 188 |
251992_at |
AT3G53350
|
MIDD1, Microtubule Depletion Domain 1, RIP3, ROP interactive partner 3, RIP4, ROP interactive partner 4 |
-0.269 |
0.042 |
| 189 |
253229_at |
AT4G34660
|
[SH3 domain-containing protein] |
-0.268 |
-0.071 |
| 190 |
257584_at |
AT1G54955
|
unknown |
-0.268 |
0.009 |
| 191 |
255062_at |
AT4G08940
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.265 |
0.103 |
| 192 |
245797_at |
AT1G45230
|
unknown |
-0.262 |
0.149 |
| 193 |
246934_at |
AT5G25290
|
unknown |
-0.261 |
0.082 |
| 194 |
262248_at |
AT1G48370
|
YSL8, YELLOW STRIPE like 8 |
-0.260 |
-0.004 |
| 195 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.255 |
-0.118 |
| 196 |
262531_at |
AT1G17230
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.254 |
-0.018 |
| 197 |
250295_at |
AT5G13390
|
NEF1, NO EXINE FORMATION 1 |
-0.252 |
0.030 |
| 198 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
-0.252 |
-0.103 |
| 199 |
264743_at |
AT1G62090
|
[pseudogene] |
-0.251 |
-0.018 |
| 200 |
248497_at |
AT5G50380
|
ATEXO70F1, exocyst subunit exo70 family protein F1, EXO70F1, exocyst subunit exo70 family protein F1 |
-0.250 |
-0.022 |
| 201 |
257855_at |
AT3G13040
|
[myb-like HTH transcriptional regulator family protein] |
-0.246 |
0.092 |
| 202 |
249531_at |
AT5G38770
|
AtGDU7, glutamine dumper 7, GDU7, glutamine dumper 7 |
-0.244 |
-0.144 |
| 203 |
263097_at |
AT2G16070
|
PDV2, PLASTID DIVISION2 |
-0.241 |
0.081 |
| 204 |
257664_at |
AT3G20400
|
EMB2743, EMBRYO DEFECTIVE 2743 |
-0.241 |
0.019 |
| 205 |
265813_at |
AT2G18060
|
ANAC037, Arabidopsis NAC domain containing protein 37, VND1, vascular related NAC-domain protein 1 |
-0.238 |
0.075 |
| 206 |
256858_at |
AT3G15140
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.237 |
0.067 |
| 207 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.235 |
-0.126 |
| 208 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.233 |
-0.051 |
| 209 |
248450_at |
AT5G51290
|
ACD5, ACCELERATED CELL DEATH 5 |
-0.232 |
-0.015 |
| 210 |
256092_at |
AT1G20696
|
HMGB3, high mobility group B3, NFD03, NFD3 |
-0.230 |
-0.027 |
| 211 |
259624_at |
AT1G43020
|
unknown |
-0.225 |
-0.123 |
| 212 |
265374_at |
AT2G06520
|
PSBX, photosystem II subunit X |
-0.225 |
0.187 |
| 213 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.225 |
-0.067 |
| 214 |
260404_at |
AT1G69950
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 1.0e-71 P-value blast match to GB:CAA29005 ORFa of Maize Ac (hAT-element) (Zea mays)] |
-0.223 |
0.161 |
| 215 |
259230_at |
AT3G07780
|
OBE1, OBERON1 |
-0.223 |
0.066 |
| 216 |
264653_at |
AT1G08980
|
AMI1, amidase 1, ATAMI1, AMIDASE-LIKE PROTEIN 1, ATTOC64-I, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I, TOC64-I, TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-I |
-0.222 |
0.252 |
| 217 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.221 |
-0.001 |
| 218 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.215 |
0.007 |
| 219 |
245946_at |
AT5G19580
|
[glyoxal oxidase-related protein] |
-0.214 |
0.122 |
| 220 |
246500_at |
AT5G16270
|
ATRAD21.3, ARABIDOPSIS HOMOLOG OF RAD21 3, SYN4, sister chromatid cohesion 1 protein 4 |
-0.213 |
-0.041 |
| 221 |
259201_at |
AT3G09080
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.212 |
0.051 |
| 222 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.211 |
0.243 |
| 223 |
265970_at |
AT2G11210
|
[Mutator-like transposase family, has a 1.1e-93 P-value blast match to Q9ZQM3 /24-192 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
-0.210 |
-0.005 |
| 224 |
262987_at |
AT1G23240
|
[Caleosin-related family protein] |
-0.210 |
0.046 |
| 225 |
245308_at |
AT4G17486
|
[PPPDE putative thiol peptidase family protein] |
-0.209 |
0.129 |
| 226 |
263119_at |
AT1G03110
|
AtTRM82, TRM82, tRNA modification 82 |
-0.209 |
0.045 |
| 227 |
254986_at |
AT4G10640
|
IQD16, IQ-domain 16 |
-0.208 |
0.092 |
| 228 |
265055_at |
AT1G52020
|
[pseudogene] |
-0.206 |
-0.005 |
| 229 |
251140_at |
AT5G01090
|
[Concanavalin A-like lectin family protein] |
-0.206 |
-0.014 |
| 230 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
-0.204 |
-0.057 |
| 231 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.204 |
-0.081 |
| 232 |
259377_at |
AT3G16380
|
PAB6, poly(A) binding protein 6 |
-0.202 |
0.100 |
| 233 |
261843_at |
AT1G16180
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-0.200 |
-0.005 |
| 234 |
256112_at |
AT1G16920
|
ATRABA1B, RAB GTPase homolog A1B, BEX5, BFA-visualized exocytic trafficking defective 5, RAB11, RABA1b, RAB GTPase homolog A1B |
-0.197 |
0.111 |
| 235 |
259376_at |
AT3G16320
|
Tetratricopeptide repeat (TPR)-like superfamily protein |
-0.196 |
0.051 |
| 236 |
248249_at |
AT5G53140
|
[Protein phosphatase 2C family protein] |
-0.195 |
-0.051 |
| 237 |
263778_at |
AT2G46470
|
OXA1L, inner membrane protein OXA1-like |
-0.195 |
0.120 |
| 238 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.194 |
-0.176 |
| 239 |
256040_at |
AT1G07270
|
[Cell division control, Cdc6] |
-0.192 |
-0.044 |