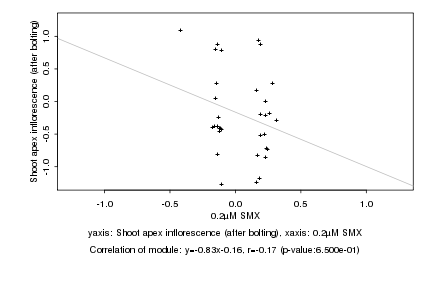
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
0.2µM SMX |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.312 |
-0.286 |
| 2 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.283 |
0.277 |
| 3 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.257 |
-0.178 |
| 4 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.244 |
-0.723 |
| 5 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.232 |
-0.708 |
| 6 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.229 |
0.006 |
| 7 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
0.227 |
-0.202 |
| 8 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
0.225 |
-0.857 |
| 9 |
251044_at |
AT5G02350
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.215 |
-0.495 |
| 10 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
0.191 |
-0.509 |
| 11 |
249019_at |
AT5G44780
|
MORF4, multiple organellar RNA editing factor 4 |
0.189 |
-0.199 |
| 12 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.186 |
0.880 |
| 13 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.178 |
-1.177 |
| 14 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.175 |
0.940 |
| 15 |
259661_at |
AT1G55265
|
unknown |
0.163 |
-0.815 |
| 16 |
249738_at |
AT5G24510
|
[60S acidic ribosomal protein family] |
0.156 |
-1.240 |
| 17 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.155 |
0.180 |
| 18 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.421 |
1.099 |
| 19 |
254132_at |
AT4G24660
|
ATHB22, HOMEOBOX PROTEIN 22, HB22, homeobox protein 22, MEE68, MATERNAL EFFECT EMBRYO ARREST 68, ZHD2, ZINC FINGER HOMEODOMAIN 2 |
-0.180 |
-0.398 |
| 20 |
255095_at |
AT4G08500
|
ARAKIN, ATMEKK1, MAPKKK8, MEKK1, MAPK/ERK kinase kinase 1 |
-0.163 |
-0.374 |
| 21 |
263907_at |
AT2G36270
|
ABI5, ABA INSENSITIVE 5, AtABI5, GIA1, GROWTH-INSENSITIVITY TO ABA 1 |
-0.159 |
0.810 |
| 22 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
-0.159 |
0.055 |
| 23 |
245401_at |
AT4G17670
|
unknown |
-0.152 |
0.279 |
| 24 |
252084_at |
AT3G51970
|
ASAT1, acyl-CoA sterol acyl transferase 1, ATASAT1, ATSAT1, ARABIDOPSIS THALIANA STEROL O-ACYLTRANSFERASE 1 |
-0.142 |
-0.809 |
| 25 |
262762_at |
AT1G10700
|
PRS3, phosphoribosyl pyrophosphate (PRPP) synthase 3 |
-0.141 |
0.886 |
| 26 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
-0.138 |
-0.382 |
| 27 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
-0.137 |
-0.230 |
| 28 |
262938_at |
AT1G79540
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.127 |
-0.405 |
| 29 |
245933_at |
AT5G09320
|
VPS9B |
-0.122 |
-0.452 |
| 30 |
266199_at |
AT2G38910
|
CPK20, calcium-dependent protein kinase 20 |
-0.116 |
-0.412 |
| 31 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.114 |
0.784 |
| 32 |
264431_at |
AT1G61700
|
[RNA polymerases N / 8 kDa subunit] |
-0.109 |
-1.258 |
| 33 |
267557_at |
AT2G32710
|
ACK2, CYCLIN-DEPENDENT KINASE INHIBITOR 2, ICK7, INTERACTORS OF CDC2 KINASE 7, KRP4, KRP4, KIP-RELATED PROTEIN 4 |
-0.107 |
-0.427 |