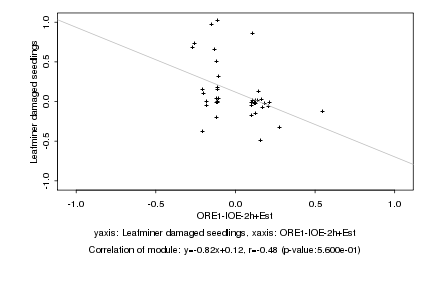
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ORE1-IOE-2h+Est |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.544 |
-0.115 |
| 2 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.273 |
-0.317 |
| 3 |
261243_at |
AT1G20180
|
unknown |
0.208 |
-0.009 |
| 4 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.203 |
-0.052 |
| 5 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.177 |
-0.023 |
| 6 |
254823_at |
AT4G12580
|
unknown |
0.165 |
-0.067 |
| 7 |
251014_at |
AT5G02520
|
KNL2, KINETOCHORE NULL 2 |
0.160 |
0.032 |
| 8 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.152 |
-0.481 |
| 9 |
244921_s_at |
ATMG01000
|
ORF114 |
0.139 |
0.135 |
| 10 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.138 |
0.015 |
| 11 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
0.125 |
-0.146 |
| 12 |
248456_at |
AT5G51380
|
[RNI-like superfamily protein] |
0.125 |
-0.021 |
| 13 |
255355_at |
AT4G03890
|
unknown |
0.123 |
0.016 |
| 14 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
0.118 |
-0.010 |
| 15 |
267210_at |
AT2G30920
|
ATCOQ3, coenzyme Q 3, COQ3, COQ3, coenzyme Q 3, EMB3002, embryo defective 3002 |
0.116 |
-0.023 |
| 16 |
254077_at |
AT4G25640
|
ATDTX35, detoxifying efflux carrier 35, DTX35, detoxifying efflux carrier 35, FFT, FLOWER FLAVONOID TRANSPORTER |
0.115 |
-0.007 |
| 17 |
267455_at |
AT2G33760
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.105 |
0.025 |
| 18 |
249541_at |
AT5G38130
|
[HXXXD-type acyl-transferase family protein] |
0.102 |
0.867 |
| 19 |
260160_at |
AT1G79880
|
AtLa2, La2, La protein 2 |
0.100 |
-0.167 |
| 20 |
262429_at |
AT1G47520
|
[copia-like retrotransposon family, has a 3.3e-256 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.099 |
-0.042 |
| 21 |
255705_at |
AT4G00160
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.099 |
-0.010 |
| 22 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.273 |
0.689 |
| 23 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.259 |
0.732 |
| 24 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.212 |
-0.371 |
| 25 |
254201_at |
AT4G24130
|
unknown |
-0.208 |
0.154 |
| 26 |
249277_at |
AT5G41890
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.204 |
0.112 |
| 27 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-0.182 |
-0.039 |
| 28 |
254566_at |
AT4G19240
|
unknown |
-0.182 |
0.009 |
| 29 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.151 |
0.976 |
| 30 |
251950_at |
AT3G53600
|
[C2H2-type zinc finger family protein] |
-0.133 |
0.663 |
| 31 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.125 |
-0.191 |
| 32 |
263332_at |
AT2G03870
|
EMB2816, EMBRYO DEFECTIVE 2816, LSM7, SM-like 7 |
-0.124 |
0.046 |
| 33 |
245842_at |
AT1G58430
|
RXF26 |
-0.124 |
-0.006 |
| 34 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.119 |
0.509 |
| 35 |
263455_at |
AT2G22320
|
unknown |
-0.117 |
0.005 |
| 36 |
266585_at |
AT2G14930
|
[copia-like retrotransposon family, has a 9.9e-255 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.117 |
-0.008 |
| 37 |
260655_at |
AT1G19320
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.114 |
0.161 |
| 38 |
246453_at |
AT5G16830
|
ATPEP12, ATSYP21, PEP12, PEP12P, SYP21, syntaxin of plants 21 |
-0.113 |
0.187 |
| 39 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
-0.113 |
1.034 |
| 40 |
262521_at |
AT1G17130
|
unknown |
-0.112 |
0.046 |
| 41 |
247596_at |
AT5G60840
|
unknown |
-0.107 |
0.324 |