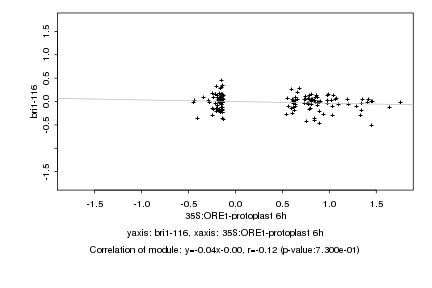
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S:ORE1-protoplast 6h |
Log2 signal ratio
bri1-116 |
| 1 |
257944_at |
AT3G21850
|
ASK9, SKP1-like 9, SK9, SKP1-like 9 |
1.754 |
-0.001 |
| 2 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
1.635 |
-0.111 |
| 3 |
257943_at |
AT3G21840
|
ASK7, SKP1-like 7, SK7, SKP1-like 7 |
1.450 |
0.014 |
| 4 |
257942_at |
AT3G21830
|
ASK8, SKP1-like 8, SK8, SKP1-like 8 |
1.449 |
0.010 |
| 5 |
254629_at |
AT4G18425
|
unknown |
1.439 |
-0.494 |
| 6 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
1.414 |
0.051 |
| 7 |
262356_at |
AT1G73000
|
PYL3, PYR1-like 3, RCAR13, regulatory components of ABA receptor 13 |
1.402 |
-0.008 |
| 8 |
261243_at |
AT1G20180
|
unknown |
1.343 |
0.063 |
| 9 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
1.341 |
-0.176 |
| 10 |
263282_at |
AT2G14095
|
unknown |
1.336 |
-0.021 |
| 11 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
1.331 |
-0.288 |
| 12 |
267343_at |
AT2G44260
|
unknown |
1.288 |
-0.102 |
| 13 |
253289_at |
AT4G34320
|
unknown |
1.200 |
-0.043 |
| 14 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
1.183 |
0.064 |
| 15 |
250535_at |
AT5G08480
|
[VQ motif-containing protein] |
1.090 |
-0.049 |
| 16 |
258960_at |
AT3G10590
|
[Duplicated homeodomain-like superfamily protein] |
1.067 |
0.071 |
| 17 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
1.064 |
0.051 |
| 18 |
248168_at |
AT5G54570
|
BGLU41, beta glucosidase 41 |
1.042 |
0.134 |
| 19 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
1.029 |
-0.281 |
| 20 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
1.027 |
-0.097 |
| 21 |
260845_at |
AT1G17310
|
[MADS-box transcription factor family protein] |
1.023 |
0.042 |
| 22 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
0.991 |
0.162 |
| 23 |
264144_at |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
0.979 |
0.028 |
| 24 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.972 |
0.136 |
| 25 |
266849_at |
AT2G25940
|
ALPHA-VPE, alpha-vacuolar processing enzyme, ALPHAVPE |
0.970 |
-0.030 |
| 26 |
247324_at |
AT5G64190
|
unknown |
0.936 |
-0.261 |
| 27 |
245705_at |
AT5G04390
|
[C2H2-type zinc finger family protein] |
0.897 |
0.009 |
| 28 |
264532_at |
AT1G55740
|
AtSIP1, seed imbibition 1, RS1, raffinose synthase 1, SIP1, seed imbibition 1 |
0.894 |
-0.196 |
| 29 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
0.892 |
-0.463 |
| 30 |
249569_at |
AT5G38070
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.886 |
-0.009 |
| 31 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.875 |
-0.005 |
| 32 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.873 |
-0.018 |
| 33 |
254956_at |
AT4G10850
|
AtSWEET7, SWEET7 |
0.869 |
0.096 |
| 34 |
250949_at |
AT5G03510
|
[C2H2-type zinc finger family protein] |
0.867 |
-0.079 |
| 35 |
257580_at |
AT3G06210
|
[ARM repeat superfamily protein] |
0.857 |
0.132 |
| 36 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.848 |
-0.008 |
| 37 |
266865_at |
AT2G29980
|
AtFAD3, FAD3, fatty acid desaturase 3 |
0.846 |
0.088 |
| 38 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.842 |
0.037 |
| 39 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.839 |
-0.360 |
| 40 |
267119_at |
AT2G32610
|
ATCSLB01, cellulose synthase-like B1, ATCSLB1, CELLULOSE SYNTHASE LIKE B1, CSLB01, CELLULOSE SYNTHASE LIKE B1, CSLB01, cellulose synthase-like B1 |
0.836 |
-0.393 |
| 41 |
256789_at |
AT3G13672
|
SINA2 |
0.816 |
0.045 |
| 42 |
265014_at |
AT1G24430
|
[HXXXD-type acyl-transferase family protein] |
0.809 |
0.043 |
| 43 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
0.808 |
-0.063 |
| 44 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.807 |
0.166 |
| 45 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.802 |
0.056 |
| 46 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.801 |
0.007 |
| 47 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
0.794 |
-0.133 |
| 48 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.787 |
0.046 |
| 49 |
245993_at |
AT5G20700
|
unknown |
0.786 |
-0.153 |
| 50 |
265539_at |
AT2G15830
|
unknown |
0.784 |
0.146 |
| 51 |
265582_at |
AT2G20030
|
[RING/U-box superfamily protein] |
0.778 |
0.067 |
| 52 |
260582_at |
AT2G47200
|
unknown |
0.774 |
-0.051 |
| 53 |
260887_at |
AT1G29160
|
[Dof-type zinc finger DNA-binding family protein] |
0.770 |
-0.029 |
| 54 |
259328_at |
AT3G16440
|
ATMLP-300B, myrosinase-binding protein-like protein-300B, MEE36, maternal effect embryo arrest 36, MLP-300B, myrosinase-binding protein-like protein-300B |
0.759 |
-0.031 |
| 55 |
249797_at |
AT5G23750
|
[Remorin family protein] |
0.751 |
-0.410 |
| 56 |
266591_at |
AT2G46225
|
ABIL1, ABI-1-like 1 |
0.743 |
0.044 |
| 57 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.738 |
0.110 |
| 58 |
252920_at |
AT4G39000
|
AtGH9B17, glycosyl hydrolase 9B17, GH9B17, glycosyl hydrolase 9B17 |
0.726 |
0.058 |
| 59 |
248761_at |
AT5G47635
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.726 |
-0.038 |
| 60 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.673 |
0.291 |
| 61 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
0.660 |
0.200 |
| 62 |
247985_at |
AT5G56790
|
[Protein kinase superfamily protein] |
0.652 |
0.045 |
| 63 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
0.635 |
-0.062 |
| 64 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.635 |
0.094 |
| 65 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.634 |
-0.089 |
| 66 |
249564_at |
AT5G38400
|
unknown |
0.630 |
0.030 |
| 67 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.628 |
-0.172 |
| 68 |
267058_at |
AT2G32510
|
MAPKKK17, mitogen-activated protein kinase kinase kinase 17 |
0.624 |
-0.009 |
| 69 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
0.618 |
0.016 |
| 70 |
262865_at |
AT1G64930
|
CYP89A7, cytochrome P450, family 87, subfamily A, polypeptide 7 |
0.614 |
0.023 |
| 71 |
262478_at |
AT1G11170
|
unknown |
0.613 |
-0.023 |
| 72 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.609 |
-0.110 |
| 73 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
0.605 |
0.052 |
| 74 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
0.599 |
-0.243 |
| 75 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
0.593 |
-0.146 |
| 76 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.591 |
0.268 |
| 77 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.562 |
-0.103 |
| 78 |
246799_at |
AT5G26940
|
DPD1, defective in pollen organelle DNA degradation1 |
0.546 |
0.082 |
| 79 |
262244_at |
AT1G48260
|
CIPK17, CBL-interacting protein kinase 17, SnRK3.21, SNF1-RELATED PROTEIN KINASE 3.21 |
0.540 |
-0.259 |
| 80 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
-0.456 |
-0.005 |
| 81 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.443 |
0.025 |
| 82 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
-0.410 |
-0.351 |
| 83 |
257950_at |
AT3G21780
|
UGT71B6, UDP-glucosyl transferase 71B6 |
-0.349 |
0.096 |
| 84 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.292 |
0.037 |
| 85 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
-0.277 |
-0.007 |
| 86 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.254 |
-0.146 |
| 87 |
256753_at |
AT3G27160
|
GHS1, GLUCOSE HYPERSENSITIVE 1 |
-0.248 |
0.186 |
| 88 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.245 |
-0.282 |
| 89 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
-0.240 |
0.095 |
| 90 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.240 |
-0.166 |
| 91 |
252032_at |
AT3G52150
|
PSRP2, plastid-specific ribosomal protein 2 |
-0.218 |
0.159 |
| 92 |
261521_at |
AT1G71830
|
ATSERK1, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1, SERK1, somatic embryogenesis receptor-like kinase 1 |
-0.218 |
-0.041 |
| 93 |
263736_at |
AT1G60000
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.213 |
0.162 |
| 94 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.211 |
0.325 |
| 95 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
-0.207 |
-0.130 |
| 96 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
-0.204 |
-0.201 |
| 97 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.200 |
0.093 |
| 98 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
-0.198 |
-0.084 |
| 99 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.196 |
-0.133 |
| 100 |
263698_at |
AT1G31100
|
[non-LTR retrotransposon family (LINE), has a 3.7e-35 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.192 |
0.143 |
| 101 |
249010_at |
AT5G44580
|
unknown |
-0.191 |
0.052 |
| 102 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.186 |
0.037 |
| 103 |
260841_at |
AT1G29195
|
unknown |
-0.185 |
-0.174 |
| 104 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
-0.182 |
-0.181 |
| 105 |
252230_at |
AT3G49810
|
[ARM repeat superfamily protein] |
-0.179 |
0.009 |
| 106 |
248205_at |
AT5G54300
|
unknown |
-0.177 |
0.095 |
| 107 |
264837_at |
AT1G03600
|
PSB27 |
-0.176 |
0.182 |
| 108 |
253665_at |
AT4G30230
|
unknown |
-0.176 |
-0.018 |
| 109 |
248706_at |
AT5G48530
|
unknown |
-0.175 |
0.038 |
| 110 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.175 |
0.084 |
| 111 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
-0.175 |
-0.157 |
| 112 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.175 |
0.083 |
| 113 |
252810_at |
AT3G42440
|
unknown |
-0.172 |
0.080 |
| 114 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.171 |
-0.197 |
| 115 |
249335_at |
AT5G41010
|
NRPB12, NRPD12, NRPE12 |
-0.171 |
-0.008 |
| 116 |
249489_at |
AT5G39090
|
[HXXXD-type acyl-transferase family protein] |
-0.168 |
-0.035 |
| 117 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.166 |
0.090 |
| 118 |
267537_at |
AT2G41880
|
AGK1, GK-1, guanylate kinase 1 |
-0.166 |
-0.013 |
| 119 |
249943_at |
AT5G22280
|
unknown |
-0.166 |
0.096 |
| 120 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
-0.162 |
-0.214 |
| 121 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
-0.161 |
0.057 |
| 122 |
257925_at |
AT3G23170
|
unknown |
-0.160 |
0.279 |
| 123 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.159 |
0.451 |
| 124 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
-0.158 |
0.164 |
| 125 |
266751_at |
AT2G47020
|
[Peptide chain release factor 1] |
-0.157 |
0.060 |
| 126 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.157 |
0.313 |
| 127 |
257583_at |
AT1G66480
|
[plastid movement impaired 2 (PMI2)] |
-0.157 |
-0.019 |
| 128 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
-0.157 |
-0.020 |
| 129 |
262788_at |
AT1G10690
|
unknown |
-0.157 |
0.012 |
| 130 |
266035_at |
AT2G05990
|
ENR1, ENOYL-ACP REDUCTASE 1, MOD1, MOSAIC DEATH 1 |
-0.155 |
0.034 |
| 131 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.154 |
-0.052 |
| 132 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.153 |
-0.186 |
| 133 |
262049_at |
AT1G80180
|
unknown |
-0.152 |
-0.047 |
| 134 |
264396_at |
AT1G12050
|
AtFAH, FAH, fumarylacetoacetate hydrolase, SSCD1, short-day sensitive cell deathshort-day sensitive cell deathshort-day sensitive cell death 111 |
-0.150 |
0.098 |
| 135 |
259193_at |
AT3G01480
|
ATCYP38, ARABIDOPSIS CYCLOPHILIN 38, CYP38, cyclophilin 38 |
-0.149 |
0.154 |
| 136 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.149 |
-0.115 |
| 137 |
254200_at |
AT4G24110
|
unknown |
-0.148 |
0.062 |
| 138 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.148 |
0.353 |
| 139 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.146 |
0.018 |
| 140 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
-0.146 |
0.114 |
| 141 |
255224_at |
AT4G05400
|
[copper ion binding] |
-0.146 |
0.059 |
| 142 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.145 |
-0.224 |
| 143 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
-0.145 |
0.000 |
| 144 |
266896_at |
AT2G45810
|
[DEA(D/H)-box RNA helicase family protein] |
-0.145 |
-0.008 |
| 145 |
266601_at |
AT2G46060
|
[transmembrane protein-related] |
-0.145 |
0.051 |
| 146 |
260055_at |
AT1G78150
|
unknown |
-0.144 |
-0.001 |
| 147 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.143 |
-0.356 |
| 148 |
255623_at |
AT4G01310
|
[Ribosomal L5P family protein] |
-0.142 |
0.158 |
| 149 |
265620_at |
AT2G27310
|
[F-box family protein] |
-0.141 |
-0.116 |
| 150 |
262946_at |
AT1G79390
|
unknown |
-0.141 |
0.064 |
| 151 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.140 |
-0.232 |
| 152 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
-0.139 |
-0.184 |
| 153 |
262544_at |
AT1G15420
|
unknown |
-0.139 |
0.085 |
| 154 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
-0.139 |
-0.155 |
| 155 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.137 |
-0.372 |
| 156 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
-0.137 |
0.059 |
| 157 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.137 |
0.134 |