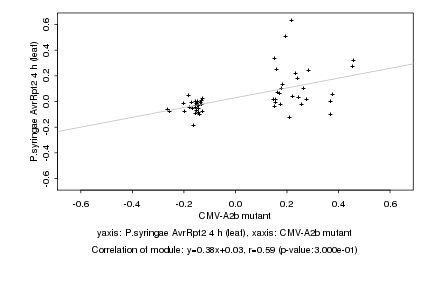
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
CMV-A2b mutant |
Log2 signal ratio
P.syringae AvrRpt2 4 h (leaf) |
| 1 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.457 |
0.321 |
| 2 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.454 |
0.278 |
| 3 |
255733_at |
AT1G25400
|
unknown |
0.376 |
0.060 |
| 4 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
0.366 |
-0.097 |
| 5 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.365 |
0.001 |
| 6 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.281 |
0.244 |
| 7 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.272 |
0.021 |
| 8 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.263 |
0.105 |
| 9 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.256 |
-0.016 |
| 10 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
0.244 |
0.037 |
| 11 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.237 |
0.180 |
| 12 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.231 |
0.221 |
| 13 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.221 |
0.042 |
| 14 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.216 |
0.639 |
| 15 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
0.208 |
-0.124 |
| 16 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.191 |
0.512 |
| 17 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.181 |
0.135 |
| 18 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
0.178 |
0.103 |
| 19 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.173 |
-0.020 |
| 20 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.170 |
0.070 |
| 21 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.162 |
0.078 |
| 22 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.157 |
0.251 |
| 23 |
250858_at |
AT5G04760
|
[Duplicated homeodomain-like superfamily protein] |
0.155 |
0.019 |
| 24 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.152 |
0.000 |
| 25 |
262085_at |
AT1G56060
|
unknown |
0.151 |
0.343 |
| 26 |
254158_at |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
0.149 |
-0.032 |
| 27 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.147 |
0.022 |
| 28 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
-0.264 |
-0.060 |
| 29 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.256 |
-0.075 |
| 30 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
-0.202 |
-0.012 |
| 31 |
262445_at |
AT1G47485
|
unknown |
-0.199 |
-0.070 |
| 32 |
259735_at |
AT1G64405
|
unknown |
-0.186 |
0.053 |
| 33 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.180 |
-0.042 |
| 34 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
-0.174 |
-0.006 |
| 35 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
-0.167 |
-0.047 |
| 36 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.165 |
-0.182 |
| 37 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.158 |
-0.045 |
| 38 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
-0.157 |
0.006 |
| 39 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
-0.155 |
-0.030 |
| 40 |
246095_at |
AT5G19310
|
CHR23, chromatin remodeling 23 |
-0.155 |
-0.089 |
| 41 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.154 |
-0.012 |
| 42 |
266507_at |
AT2G47860
|
SETH6 |
-0.153 |
-0.064 |
| 43 |
256389_at |
AT3G06220
|
[AP2/B3-like transcriptional factor family protein] |
-0.151 |
-0.024 |
| 44 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.148 |
0.007 |
| 45 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
-0.147 |
-0.084 |
| 46 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
-0.145 |
-0.026 |
| 47 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
-0.144 |
-0.049 |
| 48 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.140 |
-0.099 |
| 49 |
263002_at |
AT1G54200
|
unknown |
-0.139 |
-0.005 |
| 50 |
253243_at |
AT4G34560
|
unknown |
-0.133 |
-0.019 |
| 51 |
247066_at |
AT5G66940
|
[Dof-type zinc finger DNA-binding family protein] |
-0.132 |
0.012 |
| 52 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.130 |
0.029 |
| 53 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.128 |
-0.071 |