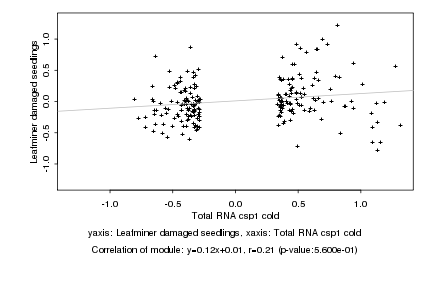
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Total RNA csp1 cold |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
1.313 |
-0.373 |
| 2 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.270 |
0.576 |
| 3 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.186 |
-0.015 |
| 4 |
246001_at |
AT5G20790
|
unknown |
1.151 |
-0.646 |
| 5 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
1.131 |
-0.780 |
| 6 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
1.126 |
-0.330 |
| 7 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
1.122 |
-0.031 |
| 8 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
1.091 |
-0.650 |
| 9 |
247474_at |
AT5G62280
|
unknown |
1.085 |
-0.400 |
| 10 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
1.084 |
-0.178 |
| 11 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
1.011 |
0.278 |
| 12 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.936 |
0.615 |
| 13 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.935 |
-0.106 |
| 14 |
246419_at |
AT5G17030
|
UGT78D3, UDP-glucosyl transferase 78D3 |
0.925 |
0.008 |
| 15 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.876 |
-0.071 |
| 16 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.868 |
-0.068 |
| 17 |
259373_at |
AT1G69160
|
unknown |
0.837 |
-0.512 |
| 18 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.829 |
0.399 |
| 19 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.806 |
1.222 |
| 20 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.794 |
0.416 |
| 21 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.759 |
0.009 |
| 22 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.753 |
0.198 |
| 23 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.729 |
0.916 |
| 24 |
252924_at |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
0.702 |
-0.002 |
| 25 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.690 |
1.009 |
| 26 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
0.684 |
-0.287 |
| 27 |
260264_at |
AT1G68500
|
unknown |
0.661 |
0.057 |
| 28 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.658 |
0.339 |
| 29 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.651 |
0.847 |
| 30 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
0.642 |
0.465 |
| 31 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.640 |
0.837 |
| 32 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
0.638 |
0.023 |
| 33 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.630 |
-0.139 |
| 34 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.629 |
0.370 |
| 35 |
260921_at |
AT1G21540
|
[AMP-dependent synthetase and ligase family protein] |
0.615 |
0.058 |
| 36 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.613 |
0.266 |
| 37 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.598 |
-0.097 |
| 38 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
0.585 |
-0.151 |
| 39 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.560 |
0.788 |
| 40 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
0.545 |
0.115 |
| 41 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
0.543 |
-0.129 |
| 42 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
0.542 |
0.220 |
| 43 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.524 |
-0.062 |
| 44 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.520 |
0.074 |
| 45 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.519 |
0.370 |
| 46 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.519 |
0.073 |
| 47 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.518 |
0.865 |
| 48 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
0.513 |
0.130 |
| 49 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.509 |
-0.048 |
| 50 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.505 |
0.448 |
| 51 |
261726_at |
AT1G76270
|
[O-fucosyltransferase family protein] |
0.494 |
-0.021 |
| 52 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.493 |
-0.712 |
| 53 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.484 |
0.915 |
| 54 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.482 |
0.155 |
| 55 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.480 |
0.040 |
| 56 |
264989_at |
AT1G27200
|
unknown |
0.480 |
0.131 |
| 57 |
263931_at |
AT2G36220
|
unknown |
0.470 |
0.605 |
| 58 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.459 |
-0.189 |
| 59 |
250053_at |
AT5G17850
|
[Sodium/calcium exchanger family protein] |
0.452 |
0.370 |
| 60 |
245533_at |
AT4G15130
|
ATCCT2, CCT2, phosphorylcholine cytidylyltransferase2 |
0.452 |
0.226 |
| 61 |
264458_at |
AT1G10410
|
unknown |
0.451 |
0.357 |
| 62 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.448 |
-0.125 |
| 63 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.447 |
0.597 |
| 64 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.443 |
0.136 |
| 65 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
0.442 |
0.209 |
| 66 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.441 |
-0.149 |
| 67 |
261089_at |
AT1G07570
|
APK1, APK1A |
0.438 |
-0.295 |
| 68 |
249411_at |
AT5G40390
|
RS5, raffinose synthase 5, SIP1, seed imbibition 1-like |
0.437 |
0.190 |
| 69 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.436 |
-0.017 |
| 70 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
0.436 |
-0.034 |
| 71 |
258509_at |
AT3G06620
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.426 |
-0.022 |
| 72 |
250974_at |
AT5G02820
|
BIN5, BRASSINOSTEROID INSENSITIVE 5, RHL2, ROOT HAIRLESS 2 |
0.426 |
0.086 |
| 73 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
0.425 |
-0.131 |
| 74 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
0.424 |
0.286 |
| 75 |
259570_at |
AT1G20440
|
AtCOR47, COR47, cold-regulated 47, RD17 |
0.419 |
0.360 |
| 76 |
252127_at |
AT3G50960
|
PLP3a, phosducin-like protein 3 homolog |
0.411 |
-0.023 |
| 77 |
259605_at |
AT1G27910
|
ATPUB45, ARABIDOPSIS THALIANA PLANT U-BOX 45, PUB45, plant U-box 45 |
0.403 |
0.117 |
| 78 |
262136_at |
AT1G77850
|
ARF17, auxin response factor 17 |
0.401 |
0.012 |
| 79 |
266049_at |
AT2G40780
|
[Nucleic acid-binding, OB-fold-like protein] |
0.392 |
0.115 |
| 80 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.387 |
-0.305 |
| 81 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.382 |
-0.048 |
| 82 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.381 |
-0.348 |
| 83 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.378 |
-0.048 |
| 84 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.376 |
0.366 |
| 85 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.373 |
0.718 |
| 86 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
0.369 |
0.005 |
| 87 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.368 |
-0.109 |
| 88 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.367 |
-0.015 |
| 89 |
266358_at |
AT2G32280
|
VCC, VASCULATURE COMPLEXITY AND CONNECTIVITY |
0.367 |
0.007 |
| 90 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
0.363 |
-0.046 |
| 91 |
262131_at |
AT1G02900
|
ATRALF1, RAPID ALKALINIZATION FACTOR 1, RALF1, rapid alkalinization factor 1, RALFL1, RALF-LIKE 1 |
0.363 |
0.350 |
| 92 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.363 |
-0.162 |
| 93 |
267631_at |
AT2G42150
|
[DNA-binding bromodomain-containing protein] |
0.362 |
0.060 |
| 94 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.362 |
-0.081 |
| 95 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.361 |
0.014 |
| 96 |
261377_at |
AT1G18850
|
unknown |
0.360 |
0.062 |
| 97 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.359 |
0.364 |
| 98 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.358 |
-0.138 |
| 99 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.358 |
-0.067 |
| 100 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
0.357 |
0.068 |
| 101 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
0.355 |
-0.227 |
| 102 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.354 |
-0.083 |
| 103 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
0.352 |
-0.061 |
| 104 |
260688_at |
AT1G17665
|
unknown |
0.349 |
0.022 |
| 105 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
0.347 |
0.399 |
| 106 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.345 |
-0.208 |
| 107 |
246536_at |
AT5G15920
|
EMB2782, EMBRYO DEFECTIVE 2782, SMC5, structural maintenance of chromosomes 5 |
0.343 |
-0.230 |
| 108 |
247878_at |
AT5G57760
|
unknown |
0.339 |
-0.372 |
| 109 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.338 |
0.127 |
| 110 |
262611_at |
AT1G14060
|
[GCK domain-containing protein] |
0.338 |
0.105 |
| 111 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.337 |
0.063 |
| 112 |
250758_at |
AT5G06000
|
ATEIF3G2, ARABIDOPSIS THALIANA EUKARYOTIC TRANSLATION INITIATION FACTOR 3G2, EIF3G2, eukaryotic translation initiation factor 3G2 |
0.335 |
-0.047 |
| 113 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.805 |
0.048 |
| 114 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.775 |
-0.261 |
| 115 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.725 |
-0.404 |
| 116 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.718 |
-0.245 |
| 117 |
252938_at |
AT4G39190
|
unknown |
-0.669 |
0.039 |
| 118 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.664 |
0.248 |
| 119 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.659 |
-0.466 |
| 120 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.659 |
0.001 |
| 121 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.650 |
-0.142 |
| 122 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.647 |
-0.203 |
| 123 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.644 |
-0.365 |
| 124 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
-0.641 |
0.727 |
| 125 |
265828_at |
AT2G14520
|
unknown |
-0.632 |
-0.131 |
| 126 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.601 |
-0.022 |
| 127 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.593 |
-0.217 |
| 128 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.584 |
-0.500 |
| 129 |
250828_at |
AT5G05250
|
unknown |
-0.578 |
-0.367 |
| 130 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.562 |
-0.107 |
| 131 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.556 |
-0.190 |
| 132 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.543 |
-0.563 |
| 133 |
253636_at |
AT4G30500
|
unknown |
-0.538 |
-0.118 |
| 134 |
259320_at |
AT3G01080
|
ATWRKY58, WRKY DNA-BINDING PROTEIN 58, WRKY58, WRKY DNA-binding protein 58 |
-0.529 |
0.225 |
| 135 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.526 |
0.487 |
| 136 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.515 |
0.006 |
| 137 |
262411_at |
AT1G34640
|
[peptidases] |
-0.508 |
-0.389 |
| 138 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.491 |
-0.263 |
| 139 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
-0.490 |
0.260 |
| 140 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.479 |
0.217 |
| 141 |
255957_at |
AT1G22160
|
unknown |
-0.467 |
-0.196 |
| 142 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.466 |
-0.007 |
| 143 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.466 |
0.318 |
| 144 |
255844_at |
AT2G33580
|
LYK5, LysM-containing receptor-like kinase 5 |
-0.465 |
0.304 |
| 145 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.457 |
-0.230 |
| 146 |
245776_at |
AT1G30260
|
unknown |
-0.444 |
0.329 |
| 147 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.444 |
0.394 |
| 148 |
258351_at |
AT3G17700
|
ATCNGC20, CYCLIC NUCLEOTIDE-GATED CHANNEL 20, CNBT1, cyclic nucleotide-binding transporter 1, CNGC20, cyclic nucleotide gated channel 20 |
-0.442 |
0.151 |
| 149 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.437 |
-0.140 |
| 150 |
261485_at |
AT1G14360
|
ATUTR3, UTR3, UDP-galactose transporter 3 |
-0.437 |
0.154 |
| 151 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.433 |
-0.313 |
| 152 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.432 |
-0.129 |
| 153 |
249377_at |
AT5G40690
|
unknown |
-0.432 |
-0.049 |
| 154 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.430 |
-0.517 |
| 155 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.429 |
-0.032 |
| 156 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.418 |
-0.388 |
| 157 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.414 |
-0.032 |
| 158 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
-0.411 |
0.022 |
| 159 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.409 |
0.187 |
| 160 |
260259_at |
AT1G74300
|
[alpha/beta-Hydrolases superfamily protein] |
-0.403 |
0.005 |
| 161 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
-0.402 |
0.213 |
| 162 |
248719_at |
AT5G47910
|
ATRBOHD, RBOHD, respiratory burst oxidase homologue D |
-0.401 |
0.171 |
| 163 |
264270_at |
AT1G60260
|
BGLU5, beta glucosidase 5 |
-0.397 |
0.049 |
| 164 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.396 |
-0.384 |
| 165 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.395 |
-0.194 |
| 166 |
265985_at |
AT2G24220
|
ATPUP5, purine permease 5, PUP5, purine permease 5 |
-0.391 |
-0.040 |
| 167 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
-0.390 |
-0.298 |
| 168 |
245826_at |
AT1G57850
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.385 |
0.031 |
| 169 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.384 |
0.490 |
| 170 |
246737_at |
AT5G27710
|
unknown |
-0.383 |
-0.096 |
| 171 |
266869_at |
AT2G44660
|
[ALG6, ALG8 glycosyltransferase family] |
-0.378 |
0.001 |
| 172 |
261221_at |
AT1G19960
|
unknown |
-0.377 |
-0.237 |
| 173 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.377 |
-0.135 |
| 174 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.375 |
-0.282 |
| 175 |
250265_at |
AT5G12900
|
unknown |
-0.373 |
-0.600 |
| 176 |
260884_at |
AT1G29240
|
unknown |
-0.369 |
-0.106 |
| 177 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.364 |
-0.210 |
| 178 |
251010_at |
AT5G02550
|
unknown |
-0.362 |
0.135 |
| 179 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.362 |
0.877 |
| 180 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.360 |
0.227 |
| 181 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.356 |
-0.233 |
| 182 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.353 |
-0.032 |
| 183 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.348 |
-0.156 |
| 184 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.346 |
0.068 |
| 185 |
256518_at |
AT1G66080
|
unknown |
-0.342 |
-0.351 |
| 186 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
-0.341 |
-0.032 |
| 187 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
-0.341 |
0.468 |
| 188 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.340 |
0.395 |
| 189 |
259545_at |
AT1G20560
|
AAE1, acyl activating enzyme 1 |
-0.340 |
-0.039 |
| 190 |
249978_at |
AT5G18850
|
unknown |
-0.339 |
-0.038 |
| 191 |
264435_at |
AT1G10360
|
ATGSTU18, glutathione S-transferase TAU 18, GST29, GLUTATHIONE S-TRANSFERASE 29, GSTU18, glutathione S-transferase TAU 18 |
-0.338 |
-0.070 |
| 192 |
259857_at |
AT1G68430
|
unknown |
-0.337 |
-0.170 |
| 193 |
250549_at |
AT5G07860
|
[HXXXD-type acyl-transferase family protein] |
-0.333 |
-0.213 |
| 194 |
245074_at |
AT2G23200
|
[Protein kinase superfamily protein] |
-0.331 |
-0.115 |
| 195 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.331 |
0.220 |
| 196 |
251493_at |
AT3G59300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.330 |
-0.141 |
| 197 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.329 |
0.283 |
| 198 |
248685_at |
AT5G48500
|
unknown |
-0.327 |
-0.145 |
| 199 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.327 |
-0.409 |
| 200 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.327 |
-0.059 |
| 201 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.325 |
0.422 |
| 202 |
266187_at |
AT2G38970
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.324 |
-0.369 |
| 203 |
257728_at |
AT3G18295
|
unknown |
-0.322 |
0.019 |
| 204 |
250960_at |
AT5G02940
|
unknown |
-0.316 |
0.256 |
| 205 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.315 |
-0.448 |
| 206 |
263135_at |
AT1G78550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.315 |
-0.024 |
| 207 |
249595_at |
AT5G37930
|
[Protein with RING/U-box and TRAF-like domains] |
-0.309 |
0.085 |
| 208 |
251759_at |
AT3G55630
|
ATDFD, DHFS-FPGS homolog D, DFD, DHFS-FPGS homolog D, FPGS3, folylpolyglutamate synthetase 3 |
-0.305 |
-0.397 |
| 209 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-0.305 |
-0.446 |
| 210 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.304 |
-0.262 |
| 211 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
-0.301 |
0.008 |
| 212 |
257600_at |
AT3G24770
|
CLE41, CLAVATA3/ESR-RELATED 41 |
-0.300 |
-0.107 |
| 213 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.300 |
0.519 |
| 214 |
252105_at |
AT3G51470
|
[Protein phosphatase 2C family protein] |
-0.297 |
-0.204 |
| 215 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
-0.295 |
0.031 |
| 216 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
-0.294 |
0.038 |
| 217 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.293 |
-0.288 |
| 218 |
245266_at |
AT4G17070
|
[peptidyl-prolyl cis-trans isomerases] |
-0.291 |
-0.001 |
| 219 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
-0.290 |
-0.414 |
| 220 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.290 |
-0.112 |
| 221 |
263987_at |
AT2G42690
|
[alpha/beta-Hydrolases superfamily protein] |
-0.289 |
-0.236 |
| 222 |
250424_at |
AT5G10550
|
GTE2, global transcription factor group E2 |
-0.287 |
-0.163 |
| 223 |
260423_at |
AT1G72470
|
ATEXO70D1, exocyst subunit exo70 family protein D1, EXO70D1, exocyst subunit exo70 family protein D1 |
-0.286 |
0.048 |