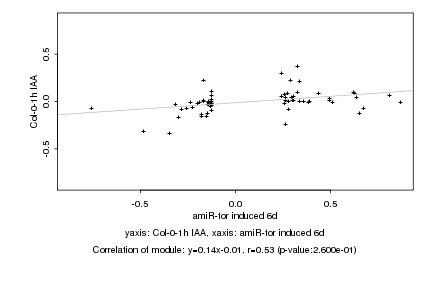
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
amiR-tor induced 6d |
Log2 signal ratio
Col-0-1h IAA |
| 1 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.866 |
-0.008 |
| 2 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.805 |
0.069 |
| 3 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.670 |
-0.067 |
| 4 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.652 |
-0.125 |
| 5 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.634 |
0.052 |
| 6 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.622 |
0.092 |
| 7 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.616 |
0.099 |
| 8 |
265539_at |
AT2G15830
|
unknown |
0.506 |
-0.002 |
| 9 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.491 |
0.034 |
| 10 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.490 |
0.021 |
| 11 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.434 |
0.089 |
| 12 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.387 |
0.008 |
| 13 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.381 |
-0.007 |
| 14 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
0.354 |
0.005 |
| 15 |
251293_at |
AT3G61930
|
unknown |
0.335 |
0.214 |
| 16 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.332 |
0.007 |
| 17 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.325 |
0.102 |
| 18 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
0.322 |
0.372 |
| 19 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.305 |
0.057 |
| 20 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.302 |
0.011 |
| 21 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.299 |
0.018 |
| 22 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.294 |
0.050 |
| 23 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.287 |
0.231 |
| 24 |
265913_at |
AT2G25625
|
unknown |
0.277 |
0.007 |
| 25 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.274 |
-0.075 |
| 26 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.271 |
0.091 |
| 27 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.261 |
0.013 |
| 28 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.260 |
0.044 |
| 29 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.259 |
-0.239 |
| 30 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.254 |
-0.011 |
| 31 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.253 |
0.083 |
| 32 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.240 |
0.056 |
| 33 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.239 |
0.301 |
| 34 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.760 |
-0.072 |
| 35 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.484 |
-0.316 |
| 36 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.351 |
-0.336 |
| 37 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.316 |
-0.021 |
| 38 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.302 |
-0.163 |
| 39 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
-0.284 |
-0.081 |
| 40 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.260 |
-0.064 |
| 41 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.237 |
-0.005 |
| 42 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
-0.229 |
-0.060 |
| 43 |
256569_at |
AT3G19550
|
unknown |
-0.201 |
-0.015 |
| 44 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
-0.190 |
-0.005 |
| 45 |
264774_at |
AT1G22890
|
unknown |
-0.182 |
-0.137 |
| 46 |
258225_at |
AT3G15630
|
unknown |
-0.179 |
-0.150 |
| 47 |
264682_at |
AT1G65570
|
[Pectin lyase-like superfamily protein] |
-0.171 |
0.225 |
| 48 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.170 |
0.013 |
| 49 |
253892_at |
AT4G27620
|
unknown |
-0.170 |
0.001 |
| 50 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.154 |
-0.150 |
| 51 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.150 |
-0.031 |
| 52 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.149 |
-0.122 |
| 53 |
264340_at |
AT1G70280
|
[NHL domain-containing protein] |
-0.144 |
0.004 |
| 54 |
264350_at |
AT1G11870
|
ATSRS, OVA7, ovule abortion 7, SRS, Seryl-tRNA synthetase |
-0.143 |
-0.025 |
| 55 |
262502_at |
AT1G21600
|
PTAC6, plastid transcriptionally active 6 |
-0.142 |
-0.001 |
| 56 |
245774_at |
AT1G30210
|
ATTCP24, TCP24, TEOSINTE BRANCHED 1, cycloidea, and PCF family 24 |
-0.136 |
-0.001 |
| 57 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.136 |
-0.045 |
| 58 |
250593_at |
AT5G07740
|
[actin binding] |
-0.131 |
-0.012 |
| 59 |
251966_at |
AT3G53380
|
LecRK-VIII.1, L-type lectin receptor kinase VIII.1 |
-0.129 |
-0.037 |
| 60 |
247078_at |
AT5G66000
|
unknown |
-0.128 |
0.026 |
| 61 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.127 |
0.073 |
| 62 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
-0.127 |
0.002 |
| 63 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.127 |
0.106 |
| 64 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.126 |
-0.093 |
| 65 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.126 |
-0.015 |