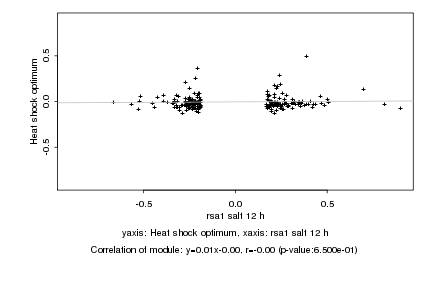
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rsa1 salt 12 h |
Log2 signal ratio
Heat shock optimum |
| 1 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
0.897 |
-0.071 |
| 2 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
0.807 |
-0.029 |
| 3 |
245692_at |
AT5G04150
|
BHLH101 |
0.693 |
0.138 |
| 4 |
256139_at |
AT1G48660
|
[Auxin-responsive GH3 family protein] |
0.504 |
-0.004 |
| 5 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.501 |
0.028 |
| 6 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
0.481 |
-0.039 |
| 7 |
260458_at |
AT1G68250
|
unknown |
0.466 |
-0.017 |
| 8 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.462 |
0.056 |
| 9 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
0.431 |
-0.030 |
| 10 |
267321_at |
AT2G19320
|
unknown |
0.420 |
-0.030 |
| 11 |
264494_at |
AT1G27461
|
unknown |
0.419 |
-0.057 |
| 12 |
263138_at |
AT1G65090
|
unknown |
0.406 |
0.011 |
| 13 |
265891_at |
AT2G15010
|
[Plant thionin] |
0.396 |
-0.027 |
| 14 |
261684_at |
AT1G47400
|
unknown |
0.387 |
0.496 |
| 15 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.382 |
-0.031 |
| 16 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
0.372 |
-0.036 |
| 17 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
0.364 |
0.003 |
| 18 |
247061_at |
AT5G66780
|
unknown |
0.356 |
-0.021 |
| 19 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.354 |
-0.044 |
| 20 |
249039_at |
AT5G44310
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.348 |
-0.007 |
| 21 |
253895_at |
AT4G27160
|
AT2S3, SESA3, seed storage albumin 3 |
0.346 |
-0.009 |
| 22 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.345 |
-0.013 |
| 23 |
257234_at |
AT3G14880
|
unknown |
0.342 |
-0.010 |
| 24 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.339 |
-0.023 |
| 25 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
0.322 |
-0.026 |
| 26 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.319 |
0.007 |
| 27 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
0.316 |
-0.017 |
| 28 |
254814_at |
AT4G12340
|
[copper ion binding] |
0.310 |
0.032 |
| 29 |
249246_at |
AT5G42290
|
[transcription activator-related] |
0.310 |
-0.014 |
| 30 |
256931_at |
AT3G22490
|
[Seed maturation protein] |
0.309 |
-0.016 |
| 31 |
259952_at |
AT1G71400
|
AtRLP12, receptor like protein 12, RLP12, receptor like protein 12 |
0.307 |
-0.076 |
| 32 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
0.301 |
-0.015 |
| 33 |
262373_at |
AT1G73120
|
unknown |
0.286 |
-0.053 |
| 34 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
0.285 |
-0.033 |
| 35 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.280 |
-0.054 |
| 36 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.275 |
-0.011 |
| 37 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.274 |
0.068 |
| 38 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
0.272 |
-0.016 |
| 39 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.271 |
-0.017 |
| 40 |
265379_at |
AT2G18340
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.267 |
-0.028 |
| 41 |
250882_at |
AT5G04000
|
unknown |
0.267 |
0.010 |
| 42 |
261169_at |
AT1G04920
|
ATSPS3F, sucrose phosphate synthase 3F, SPS3F, sucrose phosphate synthase 3F |
0.262 |
0.024 |
| 43 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
0.261 |
-0.084 |
| 44 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.255 |
0.094 |
| 45 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.254 |
-0.078 |
| 46 |
257130_at |
AT3G20210
|
DELTA-VPE, delta vacuolar processing enzyme, DELTAVPE, DELTA VACUOLAR PROCESSING ENZYME |
0.252 |
-0.027 |
| 47 |
247269_at |
AT5G64210
|
AOX2, alternative oxidase 2 |
0.246 |
-0.045 |
| 48 |
245335_at |
AT4G16160
|
ATOEP16-2, ATOEP16-S |
0.243 |
-0.026 |
| 49 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.242 |
-0.026 |
| 50 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.241 |
-0.068 |
| 51 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.241 |
0.193 |
| 52 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.236 |
0.294 |
| 53 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.235 |
0.043 |
| 54 |
259298_at |
AT3G05370
|
AtRLP31, receptor like protein 31, RLP31, receptor like protein 31 |
0.230 |
-0.026 |
| 55 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.229 |
-0.014 |
| 56 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.229 |
-0.034 |
| 57 |
257186_at |
AT3G13130
|
unknown |
0.228 |
-0.009 |
| 58 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.227 |
-0.001 |
| 59 |
258257_at |
AT3G26770
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.226 |
-0.015 |
| 60 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.226 |
-0.039 |
| 61 |
260917_at |
AT1G02700
|
unknown |
0.226 |
-0.054 |
| 62 |
250624_at |
AT5G07330
|
unknown |
0.226 |
-0.050 |
| 63 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.225 |
0.168 |
| 64 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.224 |
-0.021 |
| 65 |
256827_at |
AT3G18570
|
[Oleosin family protein] |
0.223 |
-0.026 |
| 66 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.222 |
0.143 |
| 67 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.221 |
-0.017 |
| 68 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.221 |
-0.093 |
| 69 |
254437_s_at |
AT3G06433
|
[pseudogene] |
0.220 |
-0.034 |
| 70 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
0.219 |
-0.038 |
| 71 |
264636_at |
AT1G65490
|
unknown |
0.217 |
-0.028 |
| 72 |
255550_at |
AT4G01970
|
AtSTS, stachyose synthase, RS4, raffinose synthase 4, STS, stachyose synthase |
0.214 |
-0.005 |
| 73 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.212 |
-0.126 |
| 74 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
0.212 |
0.051 |
| 75 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.210 |
-0.032 |
| 76 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.210 |
-0.001 |
| 77 |
250828_at |
AT5G05250
|
unknown |
0.210 |
0.181 |
| 78 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.209 |
0.080 |
| 79 |
259353_at |
AT3G05190
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.207 |
-0.046 |
| 80 |
259578_at |
AT1G27990
|
unknown |
0.204 |
-0.045 |
| 81 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.201 |
-0.009 |
| 82 |
253494_at |
AT4G31830
|
unknown |
0.199 |
-0.008 |
| 83 |
256898_at |
AT3G24650
|
ABI3, ABA INSENSITIVE 3, ABI3, ABSCISIC ACID INSENSITIVE 3, SIS10, SUGAR INSENSITIVE 10 |
0.197 |
-0.039 |
| 84 |
257244_at |
AT3G24240
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
0.194 |
-0.099 |
| 85 |
257421_at |
AT1G12030
|
unknown |
0.194 |
-0.028 |
| 86 |
248969_at |
AT5G45310
|
unknown |
0.194 |
0.008 |
| 87 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.194 |
-0.021 |
| 88 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
0.193 |
-0.013 |
| 89 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
0.192 |
-0.028 |
| 90 |
259042_at |
AT3G03450
|
RGL2, RGA-like 2 |
0.191 |
-0.082 |
| 91 |
263881_at |
AT2G21820
|
unknown |
0.189 |
0.010 |
| 92 |
266727_at |
AT2G03150
|
emb1579, embryo defective 1579 |
0.188 |
-0.039 |
| 93 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.186 |
-0.026 |
| 94 |
249772_at |
AT5G24130
|
unknown |
0.185 |
-0.061 |
| 95 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.184 |
0.011 |
| 96 |
261460_at |
AT1G07880
|
ATMPK13 |
0.181 |
0.070 |
| 97 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.180 |
0.013 |
| 98 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.178 |
0.061 |
| 99 |
249206_at |
AT5G42630
|
ATS, ABERRANT TESTA SHAPE, KAN4, KANADI 4 |
0.175 |
-0.046 |
| 100 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
0.173 |
-0.047 |
| 101 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.173 |
0.111 |
| 102 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.173 |
0.082 |
| 103 |
263032_at |
AT1G23850
|
unknown |
0.170 |
0.023 |
| 104 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.170 |
-0.074 |
| 105 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
0.169 |
-0.054 |
| 106 |
253154_at |
AT4G35710
|
unknown |
0.168 |
-0.030 |
| 107 |
262897_at |
AT1G59840
|
CCB4, cofactor assembly of complex C |
-0.666 |
-0.009 |
| 108 |
252934_at |
AT4G39120
|
HISN7, HISTIDINE BIOSYNTHESIS 7, IMPL2, myo-inositol monophosphatase like 2 |
-0.567 |
-0.024 |
| 109 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.531 |
-0.083 |
| 110 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.524 |
0.011 |
| 111 |
246196_at |
AT4G37090
|
unknown |
-0.522 |
0.062 |
| 112 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.457 |
-0.013 |
| 113 |
245953_at |
AT5G28520
|
[Mannose-binding lectin superfamily protein] |
-0.442 |
-0.062 |
| 114 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.425 |
0.048 |
| 115 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.394 |
0.002 |
| 116 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
-0.393 |
0.068 |
| 117 |
262066_at |
AT1G79950
|
[RAD3-like DNA-binding helicase protein] |
-0.372 |
-0.004 |
| 118 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.348 |
-0.013 |
| 119 |
245556_at |
AT4G15400
|
ABS1, ABNORMAL SHOOT 1, BIA1, BRASSINOSTEROID INACTIVATOR1 |
-0.341 |
-0.018 |
| 120 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.336 |
-0.057 |
| 121 |
246412_at |
AT5G17530
|
[phosphoglucosamine mutase family protein] |
-0.335 |
0.011 |
| 122 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.334 |
0.033 |
| 123 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.333 |
-0.001 |
| 124 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.326 |
0.068 |
| 125 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.325 |
-0.061 |
| 126 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.322 |
-0.038 |
| 127 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.320 |
-0.057 |
| 128 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
-0.316 |
0.008 |
| 129 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.312 |
0.065 |
| 130 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.308 |
-0.059 |
| 131 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
-0.305 |
-0.092 |
| 132 |
255436_at |
AT4G03150
|
unknown |
-0.298 |
-0.037 |
| 133 |
252568_at |
AT3G45410
|
LecRK-I.3, L-type lectin receptor kinase I.3 |
-0.293 |
-0.042 |
| 134 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.289 |
-0.125 |
| 135 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
-0.280 |
-0.039 |
| 136 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
-0.275 |
0.038 |
| 137 |
249108_at |
AT5G43690
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.275 |
0.002 |
| 138 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.274 |
0.210 |
| 139 |
264184_at |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.274 |
-0.006 |
| 140 |
261878_at |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
-0.273 |
-0.028 |
| 141 |
266191_at |
AT2G39040
|
[Peroxidase superfamily protein] |
-0.272 |
-0.095 |
| 142 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
-0.270 |
-0.029 |
| 143 |
253985_at |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
-0.267 |
-0.050 |
| 144 |
246888_at |
AT5G26270
|
unknown |
-0.266 |
-0.040 |
| 145 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
-0.265 |
-0.013 |
| 146 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
-0.263 |
-0.024 |
| 147 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.261 |
-0.036 |
| 148 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.260 |
-0.070 |
| 149 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.259 |
0.004 |
| 150 |
263208_at |
AT1G10480
|
ZFP5, zinc finger protein 5 |
-0.259 |
0.005 |
| 151 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.257 |
0.027 |
| 152 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
-0.253 |
-0.073 |
| 153 |
245806_at |
AT1G45474
|
Lhca5, photosystem I light harvesting complex gene 5 |
-0.252 |
-0.034 |
| 154 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.252 |
0.147 |
| 155 |
262454_at |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
-0.251 |
0.054 |
| 156 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.251 |
-0.060 |
| 157 |
245713_at |
AT5G04370
|
NAMT1 |
-0.251 |
-0.028 |
| 158 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
-0.248 |
0.024 |
| 159 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
-0.247 |
0.013 |
| 160 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.246 |
-0.023 |
| 161 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.244 |
0.018 |
| 162 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.238 |
-0.045 |
| 163 |
266900_at |
AT2G34610
|
unknown |
-0.237 |
-0.002 |
| 164 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.237 |
-0.012 |
| 165 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.235 |
0.024 |
| 166 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.234 |
-0.087 |
| 167 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
-0.234 |
-0.059 |
| 168 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.233 |
-0.003 |
| 169 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.232 |
-0.034 |
| 170 |
265021_at |
AT1G24610
|
[Rubisco methyltransferase family protein] |
-0.232 |
-0.015 |
| 171 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.228 |
0.090 |
| 172 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
-0.227 |
-0.071 |
| 173 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
-0.226 |
0.002 |
| 174 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.222 |
0.005 |
| 175 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
-0.222 |
-0.025 |
| 176 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.221 |
-0.071 |
| 177 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.221 |
0.008 |
| 178 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.220 |
0.011 |
| 179 |
264643_at |
AT1G08990
|
GUX5, Glucuronic Acid Substitution of Xylan 5, PGSIP5, plant glycogenin-like starch initiation protein 5 |
-0.218 |
-0.029 |
| 180 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.218 |
-0.068 |
| 181 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.218 |
0.258 |
| 182 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.218 |
0.010 |
| 183 |
267509_at |
AT2G45660
|
AGL20, AGAMOUS-like 20, ATSOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1, SOC1, SUPPRESSOR OF OVEREXPRESSION OF CO 1 |
-0.214 |
-0.106 |
| 184 |
246281_at |
AT4G36940
|
NAPRT1, nicotinate phosphoribosyltransferase 1 |
-0.214 |
0.000 |
| 185 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.212 |
0.052 |
| 186 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.211 |
0.363 |
| 187 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.211 |
-0.007 |
| 188 |
259296_at |
AT3G05350
|
[Metallopeptidase M24 family protein] |
-0.209 |
-0.031 |
| 189 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
-0.209 |
-0.018 |
| 190 |
267178_at |
AT2G37750
|
unknown |
-0.209 |
0.067 |
| 191 |
247511_at |
AT5G62040
|
BFT, brother of FT and TFL1 |
-0.207 |
-0.005 |
| 192 |
265014_at |
AT1G24430
|
[HXXXD-type acyl-transferase family protein] |
-0.207 |
-0.046 |
| 193 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.205 |
-0.078 |
| 194 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.203 |
-0.075 |
| 195 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.203 |
0.089 |
| 196 |
247734_at |
AT5G59400
|
unknown |
-0.202 |
-0.120 |
| 197 |
260333_at |
AT1G70500
|
[Pectin lyase-like superfamily protein] |
-0.202 |
-0.009 |
| 198 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.201 |
-0.072 |
| 199 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
-0.201 |
-0.001 |
| 200 |
249377_at |
AT5G40690
|
unknown |
-0.201 |
-0.031 |
| 201 |
266355_at |
AT2G01400
|
unknown |
-0.201 |
0.045 |
| 202 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.196 |
0.018 |
| 203 |
247634_at |
AT5G60520
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.196 |
-0.022 |
| 204 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.196 |
0.088 |
| 205 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
-0.195 |
-0.035 |
| 206 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.193 |
0.032 |
| 207 |
247832_at |
AT5G58550
|
EOL2, ETO1-like 2 |
-0.193 |
-0.050 |
| 208 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
-0.192 |
-0.004 |
| 209 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.192 |
0.018 |
| 210 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.192 |
-0.038 |
| 211 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.191 |
-0.062 |