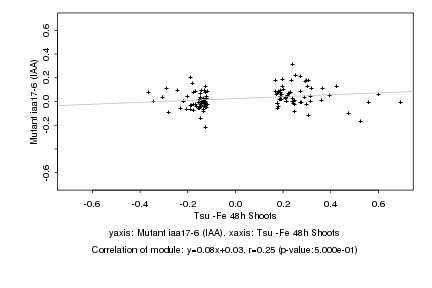
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Tsu -Fe 48h Shoots |
Log2 signal ratio
Mutant iaa17-6 (IAA) |
| 1 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.691 |
-0.006 |
| 2 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.600 |
0.066 |
| 3 |
245692_at |
AT5G04150
|
BHLH101 |
0.555 |
-0.002 |
| 4 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.524 |
-0.164 |
| 5 |
261684_at |
AT1G47400
|
unknown |
0.472 |
-0.096 |
| 6 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.420 |
0.135 |
| 7 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.393 |
0.057 |
| 8 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.363 |
0.115 |
| 9 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.361 |
0.016 |
| 10 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.317 |
0.118 |
| 11 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.313 |
0.045 |
| 12 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.311 |
0.001 |
| 13 |
250828_at |
AT5G05250
|
unknown |
0.304 |
-0.111 |
| 14 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.303 |
0.184 |
| 15 |
246125_at |
AT5G19875
|
unknown |
0.302 |
0.127 |
| 16 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.298 |
0.178 |
| 17 |
245622_at |
AT4G14080
|
MEE48, maternal effect embryo arrest 48 |
0.297 |
-0.021 |
| 18 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.291 |
0.176 |
| 19 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.286 |
0.035 |
| 20 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.277 |
-0.005 |
| 21 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.273 |
0.000 |
| 22 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.270 |
0.090 |
| 23 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.269 |
0.218 |
| 24 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.248 |
0.226 |
| 25 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.247 |
0.016 |
| 26 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.245 |
-0.081 |
| 27 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.244 |
-0.025 |
| 28 |
258362_at |
AT3G14280
|
unknown |
0.243 |
-0.012 |
| 29 |
247001_at |
AT5G67330
|
ATNRAMP4, ARABIDOPSIS THALIANA NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN 4, NRAMP4, natural resistance associated macrophage protein 4 |
0.241 |
0.003 |
| 30 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
0.240 |
0.007 |
| 31 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.238 |
0.001 |
| 32 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.238 |
0.029 |
| 33 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.236 |
0.319 |
| 34 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.235 |
0.181 |
| 35 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.229 |
0.084 |
| 36 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
0.223 |
0.069 |
| 37 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.219 |
0.068 |
| 38 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.217 |
0.056 |
| 39 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.216 |
0.046 |
| 40 |
260171_at |
AT1G71910
|
unknown |
0.212 |
0.008 |
| 41 |
256891_at |
AT3G19030
|
unknown |
0.211 |
0.002 |
| 42 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.209 |
0.032 |
| 43 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.209 |
0.041 |
| 44 |
258203_at |
AT3G13950
|
unknown |
0.199 |
0.109 |
| 45 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.197 |
0.188 |
| 46 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.195 |
0.131 |
| 47 |
260668_at |
AT1G19530
|
unknown |
0.192 |
0.020 |
| 48 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.191 |
0.065 |
| 49 |
252572_at |
AT3G45290
|
ATMLO3, MILDEW RESISTANCE LOCUS O 3, MLO3, MILDEW RESISTANCE LOCUS O 3 |
0.189 |
0.022 |
| 50 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
0.188 |
0.077 |
| 51 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.187 |
0.095 |
| 52 |
257466_at |
AT1G62840
|
unknown |
0.187 |
0.043 |
| 53 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.181 |
0.023 |
| 54 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.180 |
-0.041 |
| 55 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.177 |
0.085 |
| 56 |
258207_at |
AT3G14050
|
AT-RSH2, RELA-SPOT HOMOLOG 2, ATRSH2, RELA/SPOT HOMOLOG 2, RSH2, RELA/SPOT homolog 2 |
0.175 |
-0.011 |
| 57 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
0.173 |
-0.052 |
| 58 |
265189_at |
AT1G23840
|
unknown |
0.173 |
0.068 |
| 59 |
264151_at |
AT1G02070
|
unknown |
0.173 |
-0.010 |
| 60 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.170 |
0.066 |
| 61 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
0.167 |
0.179 |
| 62 |
258201_at |
AT3G13910
|
unknown |
0.165 |
0.090 |
| 63 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.365 |
0.077 |
| 64 |
248348_at |
AT5G52190
|
[Sugar isomerase (SIS) family protein] |
-0.345 |
0.003 |
| 65 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.310 |
0.035 |
| 66 |
247878_at |
AT5G57760
|
unknown |
-0.292 |
0.113 |
| 67 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.281 |
-0.085 |
| 68 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.244 |
0.099 |
| 69 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.231 |
-0.054 |
| 70 |
259962_at |
AT1G53690
|
[DNA directed RNA polymerase, 7 kDa subunit] |
-0.221 |
0.005 |
| 71 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.208 |
-0.060 |
| 72 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.204 |
0.050 |
| 73 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.192 |
-0.027 |
| 74 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.191 |
-0.063 |
| 75 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.189 |
0.205 |
| 76 |
252962_at |
AT4G38780
|
[Pre-mRNA-processing-splicing factor] |
-0.186 |
-0.026 |
| 77 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.181 |
0.160 |
| 78 |
249436_at |
AT5G39980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.180 |
-0.024 |
| 79 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.179 |
-0.069 |
| 80 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.179 |
0.079 |
| 81 |
258703_at |
AT3G09750
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.170 |
-0.018 |
| 82 |
267146_at |
AT2G38160
|
unknown |
-0.168 |
0.085 |
| 83 |
263953_at |
AT2G36050
|
ATOFP15, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 15, OFP15, ovate family protein 15 |
-0.166 |
-0.038 |
| 84 |
263831_at |
AT2G40300
|
ATFER4, ferritin 4, FER4, ferritin 4 |
-0.158 |
-0.003 |
| 85 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.157 |
-0.053 |
| 86 |
253599_at |
AT4G30860
|
ASHR3, ASH1-related 3, SDG4, SET domain group 4 |
-0.154 |
0.020 |
| 87 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.153 |
-0.053 |
| 88 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
-0.152 |
-0.050 |
| 89 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
-0.150 |
-0.141 |
| 90 |
263840_at |
AT2G36885
|
unknown |
-0.149 |
-0.019 |
| 91 |
247734_at |
AT5G59400
|
unknown |
-0.149 |
0.039 |
| 92 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.147 |
0.071 |
| 93 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.146 |
0.008 |
| 94 |
258959_at |
AT3G10600
|
CAT7, cationic amino acid transporter 7 |
-0.145 |
-0.038 |
| 95 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.145 |
0.096 |
| 96 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
-0.144 |
-0.015 |
| 97 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.144 |
-0.028 |
| 98 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.139 |
-0.008 |
| 99 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
-0.136 |
-0.064 |
| 100 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.136 |
-0.001 |
| 101 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.136 |
0.031 |
| 102 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.136 |
-0.081 |
| 103 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.135 |
0.011 |
| 104 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.134 |
0.097 |
| 105 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.132 |
-0.015 |
| 106 |
264369_at |
AT1G70430
|
[Protein kinase superfamily protein] |
-0.131 |
-0.002 |
| 107 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.129 |
-0.038 |
| 108 |
251485_at |
AT3G59550
|
ATRAD21.2, SISTER CHROMATID COHESION 1 PROTEIN 3, ATSYN3, SYN3 |
-0.128 |
-0.029 |
| 109 |
251981_at |
AT3G53080
|
[D-galactoside/L-rhamnose binding SUEL lectin protein] |
-0.128 |
0.005 |
| 110 |
259480_at |
AT1G19010
|
unknown |
-0.128 |
0.131 |
| 111 |
253305_at |
AT4G33666
|
unknown |
-0.128 |
0.077 |
| 112 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.128 |
-0.216 |
| 113 |
248491_at |
AT5G51010
|
[Rubredoxin-like superfamily protein] |
-0.126 |
-0.018 |
| 114 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.126 |
0.018 |
| 115 |
257666_at |
AT3G20270
|
[lipid-binding serum glycoprotein family protein] |
-0.126 |
-0.048 |
| 116 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.125 |
0.045 |
| 117 |
254762_at |
AT4G13230
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.124 |
-0.043 |
| 118 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.124 |
-0.022 |
| 119 |
258748_at |
AT3G05930
|
GLP8, germin-like protein 8 |
-0.122 |
0.027 |
| 120 |
254603_at |
AT4G19045
|
[Mob1/phocein family protein] |
-0.122 |
0.028 |
| 121 |
266035_at |
AT2G05990
|
ENR1, ENOYL-ACP REDUCTASE 1, MOD1, MOSAIC DEATH 1 |
-0.122 |
-0.020 |
| 122 |
252140_at |
AT3G51070
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.121 |
-0.020 |
| 123 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
-0.121 |
0.089 |