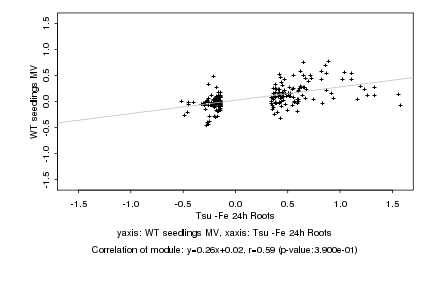
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Tsu -Fe 24h Roots |
Log2 signal ratio
WT seedlings MV |
| 1 |
260473_at |
AT1G10880
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
1.574 |
-0.062 |
| 2 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.550 |
0.149 |
| 3 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
1.326 |
0.283 |
| 4 |
251871_at |
AT3G54520
|
unknown |
1.323 |
0.128 |
| 5 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.261 |
0.127 |
| 6 |
263515_at |
AT2G21640
|
unknown |
1.232 |
0.234 |
| 7 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
1.186 |
0.289 |
| 8 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
1.166 |
0.047 |
| 9 |
250464_at |
AT5G10040
|
unknown |
1.106 |
0.548 |
| 10 |
260522_x_at |
AT2G41730
|
unknown |
1.102 |
0.426 |
| 11 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
1.040 |
0.576 |
| 12 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
1.016 |
0.437 |
| 13 |
253305_at |
AT4G33666
|
unknown |
0.932 |
0.061 |
| 14 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.910 |
0.158 |
| 15 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.886 |
0.776 |
| 16 |
250515_at |
AT5G09570
|
[Cox19-like CHCH family protein] |
0.870 |
0.225 |
| 17 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.862 |
0.551 |
| 18 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.858 |
0.702 |
| 19 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.831 |
-0.019 |
| 20 |
245226_at |
AT3G29970
|
[B12D protein] |
0.822 |
0.428 |
| 21 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.814 |
0.586 |
| 22 |
260755_at |
AT1G48980
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.743 |
0.056 |
| 23 |
246584_at |
AT5G14730
|
unknown |
0.718 |
0.446 |
| 24 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.709 |
0.518 |
| 25 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.694 |
0.402 |
| 26 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.676 |
0.231 |
| 27 |
252946_at |
AT4G39235
|
unknown |
0.662 |
0.077 |
| 28 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.661 |
0.461 |
| 29 |
253796_at |
AT4G28460
|
unknown |
0.653 |
0.274 |
| 30 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
0.649 |
0.281 |
| 31 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.647 |
0.505 |
| 32 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.647 |
0.755 |
| 33 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.635 |
0.123 |
| 34 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.629 |
0.288 |
| 35 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
0.621 |
0.588 |
| 36 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.613 |
0.300 |
| 37 |
254200_at |
AT4G24110
|
unknown |
0.603 |
0.254 |
| 38 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.601 |
0.213 |
| 39 |
265659_at |
AT2G25440
|
AtRLP20, receptor like protein 20, RLP20, receptor like protein 20 |
0.601 |
0.005 |
| 40 |
265192_at |
AT1G05060
|
unknown |
0.598 |
0.040 |
| 41 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
0.597 |
0.208 |
| 42 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.592 |
-0.191 |
| 43 |
266618_at |
AT2G35480
|
unknown |
0.591 |
0.056 |
| 44 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
0.588 |
-0.021 |
| 45 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.564 |
-0.006 |
| 46 |
251755_at |
AT3G55790
|
unknown |
0.557 |
0.002 |
| 47 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.551 |
0.505 |
| 48 |
266571_at |
AT2G23830
|
[PapD-like superfamily protein] |
0.551 |
0.093 |
| 49 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.547 |
0.264 |
| 50 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.539 |
0.233 |
| 51 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.538 |
-0.062 |
| 52 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.525 |
0.158 |
| 53 |
251761_at |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
0.521 |
0.105 |
| 54 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.508 |
0.111 |
| 55 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.508 |
0.280 |
| 56 |
254978_at |
AT4G10540
|
[Subtilase family protein] |
0.501 |
0.124 |
| 57 |
254055_at |
AT4G25330
|
unknown |
0.500 |
0.161 |
| 58 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.500 |
0.154 |
| 59 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.489 |
-0.162 |
| 60 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
0.486 |
0.112 |
| 61 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.473 |
0.169 |
| 62 |
248370_at |
AT5G52170
|
HDG7, homeodomain GLABROUS 7 |
0.470 |
-0.042 |
| 63 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.465 |
0.220 |
| 64 |
249384_at |
AT5G39890
|
unknown |
0.462 |
0.436 |
| 65 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.456 |
0.164 |
| 66 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.455 |
0.115 |
| 67 |
262336_at |
AT1G64220
|
TOM7-2, translocase of outer membrane 7 kDa subunit 2 |
0.450 |
0.022 |
| 68 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.447 |
0.107 |
| 69 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.444 |
0.092 |
| 70 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.443 |
0.185 |
| 71 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.442 |
0.311 |
| 72 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.440 |
-0.087 |
| 73 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
0.437 |
0.369 |
| 74 |
267110_at |
AT2G14800
|
unknown |
0.435 |
0.012 |
| 75 |
245324_at |
AT4G17260
|
[Lactate/malate dehydrogenase family protein] |
0.429 |
0.098 |
| 76 |
250152_at |
AT5G15120
|
unknown |
0.428 |
0.462 |
| 77 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.426 |
0.066 |
| 78 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.424 |
-0.310 |
| 79 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.417 |
0.123 |
| 80 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.416 |
0.153 |
| 81 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.416 |
0.174 |
| 82 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.415 |
-0.027 |
| 83 |
247024_at |
AT5G66985
|
unknown |
0.415 |
0.536 |
| 84 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.414 |
0.249 |
| 85 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.412 |
-0.013 |
| 86 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.407 |
0.233 |
| 87 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.405 |
0.171 |
| 88 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.405 |
0.108 |
| 89 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.398 |
-0.207 |
| 90 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.392 |
0.235 |
| 91 |
254419_at |
AT4G21490
|
NDB3, NAD(P)H dehydrogenase B3 |
0.388 |
-0.029 |
| 92 |
250488_at |
AT5G09700
|
[Glycosyl hydrolase family protein] |
0.380 |
0.111 |
| 93 |
246761_at |
AT5G27980
|
[Seed maturation protein] |
0.380 |
-0.015 |
| 94 |
252864_at |
AT4G39740
|
HCC2, homologue of copper chaperone SCO1 2 |
0.380 |
-0.032 |
| 95 |
262213_at |
AT1G74870
|
[RING/U-box superfamily protein] |
0.378 |
0.327 |
| 96 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.376 |
0.185 |
| 97 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.375 |
0.078 |
| 98 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.374 |
0.254 |
| 99 |
253725_at |
AT4G29340
|
PRF4, profilin 4 |
0.372 |
0.112 |
| 100 |
248028_at |
AT5G55620
|
unknown |
0.366 |
-0.239 |
| 101 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
0.363 |
0.042 |
| 102 |
255075_at |
AT4G09110
|
[RING/U-box superfamily protein] |
0.362 |
0.154 |
| 103 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.362 |
0.268 |
| 104 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.357 |
-0.096 |
| 105 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
0.354 |
-0.151 |
| 106 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.348 |
0.068 |
| 107 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.347 |
-0.058 |
| 108 |
266109_at |
AT2G37890
|
[Mitochondrial substrate carrier family protein] |
0.343 |
0.018 |
| 109 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.342 |
0.088 |
| 110 |
248086_at |
AT5G55490
|
ATGEX1, GAMETE EXPRESSED PROTEIN 1, GEX1, gamete expressed protein 1 |
0.341 |
-0.036 |
| 111 |
246872_at |
AT5G26080
|
[proline-rich family protein] |
-0.517 |
0.018 |
| 112 |
263552_x_at |
AT2G24980
|
EXT6, extensin 6 |
-0.492 |
-0.251 |
| 113 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.465 |
-0.209 |
| 114 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.458 |
-0.045 |
| 115 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.450 |
-0.006 |
| 116 |
256811_at |
AT3G21340
|
[Leucine-rich repeat protein kinase family protein] |
-0.407 |
-0.017 |
| 117 |
256283_at |
AT3G12540
|
unknown |
-0.333 |
-0.057 |
| 118 |
257832_at |
AT3G26740
|
CCL, CCR-like |
-0.328 |
-0.054 |
| 119 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
-0.314 |
-0.020 |
| 120 |
253353_at |
AT4G33730
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.303 |
-0.046 |
| 121 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.298 |
-0.067 |
| 122 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.295 |
-0.144 |
| 123 |
265480_at |
AT2G15970
|
ATCOR413-PM1, ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1, ATCYP19, CYCLOPHILLIN 19, COR413-PM1, cold regulated 413 plasma membrane 1, FL3-5A3, WCOR413, WCOR413-LIKE |
-0.293 |
-0.056 |
| 124 |
247320_at |
AT5G64040
|
PSAN |
-0.293 |
-0.011 |
| 125 |
252987_at |
AT4G38390
|
RHS17, root hair specific 17 |
-0.279 |
0.009 |
| 126 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.277 |
-0.443 |
| 127 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
-0.276 |
0.004 |
| 128 |
252517_at |
AT3G46340
|
[Leucine-rich repeat protein kinase family protein] |
-0.275 |
0.028 |
| 129 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.272 |
-0.391 |
| 130 |
264375_at |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-0.265 |
-0.011 |
| 131 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.265 |
0.328 |
| 132 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
-0.261 |
-0.065 |
| 133 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.261 |
-0.435 |
| 134 |
245495_at |
AT4G16400
|
unknown |
-0.258 |
0.061 |
| 135 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.250 |
-0.370 |
| 136 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.249 |
-0.280 |
| 137 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
-0.245 |
-0.073 |
| 138 |
262858_at |
AT1G14940
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.238 |
0.133 |
| 139 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
-0.235 |
-0.066 |
| 140 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
-0.231 |
-0.003 |
| 141 |
252236_at |
AT3G49930
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.218 |
0.044 |
| 142 |
267045_at |
AT2G34180
|
ATWL2, WPL4-LIKE 2, CIPK13, CBL-interacting protein kinase 13, SnRK3.7, SNF1-RELATED PROTEIN KINASE 3.7, WL2, WPL4-LIKE 2 |
-0.216 |
0.483 |
| 143 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
-0.215 |
-0.015 |
| 144 |
266583_at |
AT2G46220
|
[Uncharacterized conserved protein (DUF2358)] |
-0.212 |
-0.041 |
| 145 |
255667_at |
AT4G00240
|
PLDBETA2, phospholipase D beta 2 |
-0.210 |
-0.079 |
| 146 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
-0.209 |
0.036 |
| 147 |
250778_at |
AT5G05500
|
MOP10 |
-0.205 |
-0.280 |
| 148 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.204 |
-0.091 |
| 149 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
-0.204 |
-0.096 |
| 150 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.204 |
-0.068 |
| 151 |
260918_at |
AT1G21510
|
unknown |
-0.203 |
0.034 |
| 152 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
-0.202 |
-0.022 |
| 153 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.201 |
-0.019 |
| 154 |
248577_at |
AT5G49870
|
[Mannose-binding lectin superfamily protein] |
-0.200 |
0.063 |
| 155 |
267435_at |
AT2G33800
|
EMB3113, EMBRYO DEFECTIVE 3113 |
-0.196 |
-0.013 |
| 156 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.195 |
-0.288 |
| 157 |
248182_at |
AT5G54030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.189 |
0.043 |
| 158 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
-0.188 |
-0.046 |
| 159 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.188 |
-0.041 |
| 160 |
263327_at |
AT2G15300
|
[Leucine-rich repeat protein kinase family protein] |
-0.187 |
0.083 |
| 161 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
-0.187 |
-0.030 |
| 162 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.186 |
-0.158 |
| 163 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.185 |
-0.007 |
| 164 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.185 |
0.273 |
| 165 |
263134_at |
AT1G78570
|
ATRHM1, ARABIDOPSIS THALIANA RHAMNOSE BIOSYNTHESIS 1, RHM1, rhamnose biosynthesis 1, ROL1, REPRESSOR OF LRX1 1 |
-0.184 |
0.036 |
| 166 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.184 |
-0.057 |
| 167 |
246417_at |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
-0.183 |
0.080 |
| 168 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.182 |
0.027 |
| 169 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.182 |
0.107 |
| 170 |
254837_at |
AT4G12360
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.180 |
0.045 |
| 171 |
267537_at |
AT2G41880
|
AGK1, GK-1, guanylate kinase 1 |
-0.180 |
0.015 |
| 172 |
251388_at |
AT3G60840
|
MAP65-4, microtubule-associated protein 65-4 |
-0.177 |
0.035 |
| 173 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
-0.176 |
-0.026 |
| 174 |
246585_at |
AT5G14750
|
ATMYB66, myb domain protein 66, MYB66, myb domain protein 66, WER, WEREWOLF, WER1, WEREWOLF 1 |
-0.174 |
-0.034 |
| 175 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
-0.173 |
-0.272 |
| 176 |
251243_at |
AT3G61870
|
unknown |
-0.173 |
-0.007 |
| 177 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.173 |
-0.173 |
| 178 |
267222_at |
AT2G43880
|
[Pectin lyase-like superfamily protein] |
-0.172 |
-0.023 |
| 179 |
257231_at |
AT3G16550
|
DEG12, degradation of periplasmic proteins 12, DEGP12, DEGP protease 12 |
-0.172 |
0.018 |
| 180 |
257480_at |
AT1G15640
|
unknown |
-0.171 |
-0.159 |
| 181 |
247356_at |
AT5G63800
|
BGAL6, beta-galactosidase 6, MUM2, MUCILAGE-MODIFIED 2 |
-0.169 |
0.048 |
| 182 |
245607_at |
AT4G14330
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.169 |
-0.082 |
| 183 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.168 |
-0.017 |
| 184 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
-0.166 |
0.179 |
| 185 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
-0.164 |
-0.184 |
| 186 |
260796_at |
AT1G78360
|
ATGSTU21, glutathione S-transferase TAU 21, GSTU21, glutathione S-transferase TAU 21 |
-0.163 |
0.033 |
| 187 |
248048_at |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-0.163 |
0.128 |
| 188 |
267402_at |
AT2G26180
|
IQD6, IQ-domain 6 |
-0.162 |
-0.016 |
| 189 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.161 |
0.004 |
| 190 |
266612_at |
AT2G14860
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.158 |
-0.087 |
| 191 |
250058_at |
AT5G17870
|
PSRP6, plastid-specific 50S ribosomal protein 6 |
-0.158 |
-0.003 |
| 192 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.157 |
-0.138 |
| 193 |
245201_at |
AT1G67840
|
CSK, chloroplast sensor kinase |
-0.157 |
-0.066 |
| 194 |
254263_at |
AT4G23493
|
unknown |
-0.157 |
0.081 |
| 195 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.154 |
-0.024 |
| 196 |
245016_at |
ATCG00500
|
ACCD, acetyl-CoA carboxylase carboxyl transferase subunit beta |
-0.153 |
0.103 |
| 197 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.152 |
-0.056 |
| 198 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.151 |
-0.044 |
| 199 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
-0.150 |
-0.088 |
| 200 |
259703_at |
AT1G77790
|
[Glycosyl hydrolase superfamily protein] |
-0.150 |
0.105 |
| 201 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.149 |
0.028 |
| 202 |
258286_at |
AT3G16060
|
[ATP binding microtubule motor family protein] |
-0.149 |
0.015 |
| 203 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.149 |
-0.062 |
| 204 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
-0.149 |
-0.090 |
| 205 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.148 |
0.093 |
| 206 |
255411_at |
AT4G03110
|
AtBRN1, AtRBP-DR1, RNA-binding protein-defense related 1, BRN1, Bruno-like 1, RBP-DR1, RNA-binding protein-defense related 1 |
-0.148 |
0.127 |
| 207 |
257710_at |
AT3G27350
|
unknown |
-0.147 |
-0.069 |
| 208 |
263183_at |
AT1G05570
|
ATGSL06, ATGSL6, CALS1, callose synthase 1, GSL06, GSL6, GLUCAN SYNTHASE-LIKE 6 |
-0.147 |
-0.078 |
| 209 |
264250_at |
AT1G78680
|
ATGGH2, gamma-glutamyl hydrolase 2, GGH2, gamma-glutamyl hydrolase 2 |
-0.147 |
0.007 |
| 210 |
256393_at |
AT3G06280
|
[F-box associated ubiquitination effector family protein] |
-0.147 |
0.186 |
| 211 |
252504_at |
AT3G46590
|
ATTRP2, TRFL1, TRF-like 1, TRP2 |
-0.146 |
-0.122 |
| 212 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
-0.146 |
0.000 |
| 213 |
251199_at |
AT3G62980
|
AtTIR1, TIR1, TRANSPORT INHIBITOR RESPONSE 1 |
-0.146 |
0.007 |
| 214 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
-0.145 |
-0.162 |
| 215 |
261944_at |
AT1G64650
|
[Major facilitator superfamily protein] |
-0.145 |
-0.027 |
| 216 |
266891_at |
AT2G44690
|
ARAC9, Arabidopsis RAC-like 9, ATROP8, RHO-RELATED PROTEIN FROM PLANTS 8, ROP8, RHO-RELATED PROTEIN FROM PLANTS 8 |
-0.145 |
0.010 |
| 217 |
255671_at |
AT4G00355
|
ATI2, ATG8-interacting protein 2 |
-0.145 |
-0.003 |
| 218 |
263491_at |
AT2G42600
|
ATPPC2, phosphoenolpyruvate carboxylase 2, PPC2, phosphoenolpyruvate carboxylase 2 |
-0.145 |
0.083 |
| 219 |
254325_at |
AT4G22650
|
unknown |
-0.144 |
-0.045 |