|
probeID |
AGICode |
Annotation |
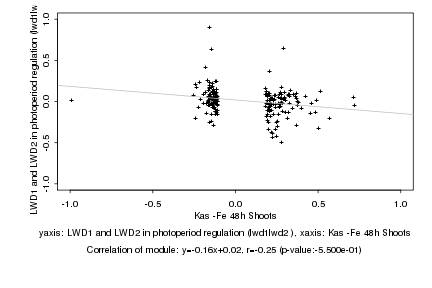
Log2 signal ratio
Kas -Fe 48h Shoots |
Log2 signal ratio
LWD1 and LWD2 in photoperiod regulation (lwd1lwd2 ) |
| 1 |
261684_at |
AT1G47400
|
unknown |
0.717 |
-0.045 |
| 2 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
0.711 |
0.051 |
| 3 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.567 |
-0.201 |
| 4 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.512 |
0.127 |
| 5 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.501 |
-0.323 |
| 6 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.485 |
0.023 |
| 7 |
245692_at |
AT5G04150
|
BHLH101 |
0.482 |
-0.123 |
| 8 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.458 |
-0.016 |
| 9 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.448 |
-0.145 |
| 10 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.418 |
0.071 |
| 11 |
250828_at |
AT5G05250
|
unknown |
0.397 |
-0.077 |
| 12 |
260349_at |
AT1G69400
|
[Transducin/WD40 repeat-like superfamily protein] |
0.376 |
-0.007 |
| 13 |
256465_at |
AT1G32570
|
unknown |
0.370 |
0.002 |
| 14 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.370 |
-0.048 |
| 15 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.369 |
-0.280 |
| 16 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.368 |
0.105 |
| 17 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.367 |
0.039 |
| 18 |
245814_at |
AT1G49910
|
BUB3.2, BUB (BUDDING UNINHIBITED BY BENZYMIDAZOL) 3.2 |
0.362 |
0.071 |
| 19 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.351 |
0.093 |
| 20 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.344 |
-0.073 |
| 21 |
264763_at |
AT1G61450
|
unknown |
0.332 |
0.139 |
| 22 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.325 |
0.069 |
| 23 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
0.325 |
-0.021 |
| 24 |
265592_at |
AT2G20110
|
[Tesmin/TSO1-like CXC domain-containing protein] |
0.322 |
0.088 |
| 25 |
257524_at |
AT3G01330
|
DEL3, DP-E2F-like protein 3, E2FF, E2L2, E2F-LIKE 2 |
0.320 |
0.090 |
| 26 |
259679_at |
AT1G77720
|
PPK1, putative protein kinase 1 |
0.318 |
0.085 |
| 27 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.315 |
-0.123 |
| 28 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
0.311 |
-0.195 |
| 29 |
260430_at |
AT1G68200
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.300 |
-0.124 |
| 30 |
254055_at |
AT4G25330
|
unknown |
0.298 |
0.017 |
| 31 |
252351_at |
AT3G48210
|
unknown |
0.296 |
0.055 |
| 32 |
259462_at |
AT1G18940
|
[Nodulin-like / Major Facilitator Superfamily protein] |
0.295 |
0.041 |
| 33 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
0.292 |
0.116 |
| 34 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.286 |
0.648 |
| 35 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.285 |
0.027 |
| 36 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.283 |
-0.016 |
| 37 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.281 |
-0.113 |
| 38 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.276 |
0.040 |
| 39 |
258405_at |
AT3G17590
|
BSH, BUSHY GROWTH, CHE1 |
0.275 |
0.048 |
| 40 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
0.275 |
0.173 |
| 41 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
0.274 |
-0.486 |
| 42 |
267410_at |
AT2G34920
|
EDA18, embryo sac development arrest 18 |
0.274 |
-0.005 |
| 43 |
250601_at |
AT5G07810
|
[SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein] |
0.273 |
0.097 |
| 44 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.272 |
-0.019 |
| 45 |
256351_at |
AT1G54960
|
ANP2, NPK1-related protein kinase 2, MAPKKK2, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASES 2, NP2, NPK1-related protein kinase 2 |
0.272 |
0.044 |
| 46 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.272 |
-0.060 |
| 47 |
264358_at |
AT1G03180
|
unknown |
0.267 |
0.120 |
| 48 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.265 |
-0.063 |
| 49 |
264623_at |
AT1G09000
|
ANP1, NPK1-related protein kinase 1, MAPKKK1, MAP KINASE KINASE KINASE 1, NP1, NPK1-related protein kinase 1 |
0.264 |
-0.034 |
| 50 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
0.263 |
-0.058 |
| 51 |
246683_at |
AT5G33300
|
[chromosome-associated kinesin-related] |
0.263 |
0.086 |
| 52 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.259 |
-0.155 |
| 53 |
254565_at |
AT4G19130
|
[Replication factor-A protein 1-related] |
0.257 |
0.052 |
| 54 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.256 |
-0.023 |
| 55 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.252 |
-0.237 |
| 56 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.251 |
-0.302 |
| 57 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.246 |
-0.252 |
| 58 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.243 |
-0.419 |
| 59 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.242 |
-0.071 |
| 60 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.239 |
-0.026 |
| 61 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.238 |
0.082 |
| 62 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.238 |
-0.330 |
| 63 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.234 |
0.025 |
| 64 |
265465_at |
AT2G37070
|
unknown |
0.232 |
0.077 |
| 65 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.226 |
-0.155 |
| 66 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.225 |
0.021 |
| 67 |
245296_at |
AT4G16370
|
oligopeptide transporter |
0.225 |
-0.027 |
| 68 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.225 |
0.023 |
| 69 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.223 |
-0.429 |
| 70 |
260522_x_at |
AT2G41730
|
unknown |
0.222 |
-0.378 |
| 71 |
267521_at |
AT2G30480
|
unknown |
0.222 |
0.039 |
| 72 |
253768_at |
AT4G28550
|
[Ypt/Rab-GAP domain of gyp1p superfamily protein] |
0.218 |
-0.106 |
| 73 |
261569_at |
AT1G01060
|
LHY, LATE ELONGATED HYPOCOTYL, LHY1, LATE ELONGATED HYPOCOTYL 1 |
0.217 |
-0.372 |
| 74 |
264964_at |
AT1G60460
|
unknown |
0.215 |
0.026 |
| 75 |
254287_at |
AT4G22960
|
unknown |
0.214 |
0.044 |
| 76 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.213 |
-0.026 |
| 77 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.212 |
-0.172 |
| 78 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.212 |
-0.018 |
| 79 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
0.210 |
0.044 |
| 80 |
247597_at |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
0.208 |
-0.059 |
| 81 |
255016_at |
AT4G10120
|
ATSPS4F |
0.206 |
-0.113 |
| 82 |
251844_at |
AT3G54630
|
unknown |
0.205 |
0.085 |
| 83 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.203 |
0.016 |
| 84 |
252165_at |
AT3G50550
|
unknown |
0.201 |
-0.085 |
| 85 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.201 |
0.369 |
| 86 |
253357_at |
AT4G33400
|
[Vacuolar import/degradation, Vid27-related protein] |
0.201 |
0.103 |
| 87 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.200 |
-0.029 |
| 88 |
261772_at |
AT1G76240
|
unknown |
0.199 |
-0.247 |
| 89 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.198 |
0.109 |
| 90 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.198 |
-0.104 |
| 91 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
0.196 |
-0.065 |
| 92 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.196 |
-0.330 |
| 93 |
245617_at |
AT4G14490
|
[SMAD/FHA domain-containing protein ] |
0.192 |
0.073 |
| 94 |
251037_at |
AT5G02100
|
ORP3A, OSBP(OXYSTEROL BINDING PROTEIN)-RELATED PROTEIN 3A, UNE18, UNFERTILIZED EMBRYO SAC 18 |
0.192 |
-0.023 |
| 95 |
257174_at |
AT3G27190
|
UKL2, uridine kinase-like 2 |
0.191 |
0.023 |
| 96 |
257545_at |
AT3G23200
|
[Uncharacterised protein family (UPF0497)] |
0.190 |
-0.149 |
| 97 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.190 |
0.063 |
| 98 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.189 |
-0.228 |
| 99 |
255434_at |
AT4G03180
|
unknown |
0.189 |
0.064 |
| 100 |
259546_at |
AT1G35350
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.189 |
-0.097 |
| 101 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.187 |
-0.178 |
| 102 |
258810_at |
AT3G03970
|
[ARM repeat superfamily protein] |
0.186 |
0.094 |
| 103 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.182 |
-0.020 |
| 104 |
255613_at |
AT4G01270
|
[RING/U-box superfamily protein] |
0.182 |
0.127 |
| 105 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.179 |
0.162 |
| 106 |
247910_at |
AT5G57410
|
[Afadin/alpha-actinin-binding protein] |
0.177 |
0.087 |
| 107 |
257086_at |
AT3G20490
|
unknown protein; Has 754 Blast hits to 165 proteins in 64 species: Archae - 0; Bacteria - 48; Metazoa - 26; Fungi - 25; Plants - 36; Viruses - 0; Other Eukaryotes - 619 (source: NCBI BLink). |
0.176 |
-0.053 |
| 108 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.996 |
0.021 |
| 109 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.254 |
0.078 |
| 110 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.245 |
-0.206 |
| 111 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.244 |
0.213 |
| 112 |
263831_at |
AT2G40300
|
ATFER4, ferritin 4, FER4, ferritin 4 |
-0.238 |
0.182 |
| 113 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
-0.225 |
-0.071 |
| 114 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
-0.219 |
0.236 |
| 115 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.215 |
0.031 |
| 116 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.198 |
-0.018 |
| 117 |
259394_at |
AT1G06420
|
unknown |
-0.194 |
0.097 |
| 118 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.184 |
0.425 |
| 119 |
262840_at |
AT1G14900
|
HMGA, high mobility group A |
-0.183 |
0.110 |
| 120 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
-0.180 |
-0.013 |
| 121 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.178 |
-0.134 |
| 122 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.175 |
0.263 |
| 123 |
251010_at |
AT5G02550
|
unknown |
-0.169 |
-0.005 |
| 124 |
255727_at |
AT1G25510
|
[Eukaryotic aspartyl protease family protein] |
-0.169 |
0.135 |
| 125 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.166 |
0.060 |
| 126 |
259183_at |
AT3G01580
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.165 |
0.072 |
| 127 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.165 |
0.022 |
| 128 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.164 |
-0.044 |
| 129 |
265908_at |
AT4G00270
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.164 |
0.005 |
| 130 |
256887_at |
AT3G15150
|
ATMMS21, A. THALIANA METHYL METHANE SULFONATE SENSITIVITY 21, HPY2, HIGH PLOIDY2, MMS21, METHYL METHANE SULFONATE SENSITIVITY 2 |
-0.162 |
0.051 |
| 131 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.160 |
0.908 |
| 132 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
-0.160 |
0.200 |
| 133 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.159 |
-0.244 |
| 134 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.159 |
0.159 |
| 135 |
256389_at |
AT3G06220
|
[AP2/B3-like transcriptional factor family protein] |
-0.159 |
0.233 |
| 136 |
245673_at |
AT1G56690
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.158 |
0.173 |
| 137 |
250245_at |
AT5G13690
|
CYL1, CYCLOPS 1, NAGLU, N-ACETYL-GLUCOSAMINIDASE |
-0.158 |
0.093 |
| 138 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.158 |
0.032 |
| 139 |
263611_at |
AT2G16450
|
[F-box and associated interaction domains-containing protein] |
-0.157 |
-0.043 |
| 140 |
251162_at |
AT3G63300
|
FKD1, FORKED 1 |
-0.155 |
0.064 |
| 141 |
252129_at |
AT3G50890
|
AtHB28, homeobox protein 28, HB28, homeobox protein 28, ZHD7, ZINC FINGER HOMEODOMAIN 7 |
-0.153 |
0.062 |
| 142 |
263899_at |
AT2G21710
|
EMB2219, embryo defective 2219 |
-0.152 |
0.097 |
| 143 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
-0.152 |
0.086 |
| 144 |
248481_at |
AT5G50930
|
[Histone superfamily protein] |
-0.151 |
0.045 |
| 145 |
257815_at |
AT3G25130
|
unknown |
-0.149 |
-0.155 |
| 146 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.149 |
0.642 |
| 147 |
259346_at |
AT3G03910
|
glutamate dehydrogenase 3 |
-0.146 |
0.060 |
| 148 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.146 |
-0.240 |
| 149 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
-0.146 |
-0.017 |
| 150 |
258027_at |
AT3G19515
|
unknown |
-0.146 |
-0.028 |
| 151 |
263485_at |
AT2G29890
|
ATVLN1, VLN1, villin 1 |
-0.146 |
0.176 |
| 152 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
-0.144 |
-0.047 |
| 153 |
262236_at |
AT1G48330
|
unknown |
-0.143 |
-0.028 |
| 154 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.143 |
0.122 |
| 155 |
257724_at |
AT3G18510
|
unknown |
-0.143 |
0.189 |
| 156 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
-0.142 |
0.028 |
| 157 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.141 |
-0.065 |
| 158 |
257696_at |
AT3G12690
|
AGC1.5, AGC kinase 1.5 |
-0.141 |
0.035 |
| 159 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.140 |
0.220 |
| 160 |
247548_at |
AT5G61400
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.139 |
0.113 |
| 161 |
249028_at |
AT5G44740
|
POLH, Y-family DNA polymerase H |
-0.138 |
-0.062 |
| 162 |
255876_at |
AT2G40480
|
unknown |
-0.138 |
0.021 |
| 163 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
-0.137 |
0.150 |
| 164 |
259457_at |
AT1G43950
|
ARF23, auxin response factor 23 |
-0.137 |
0.014 |
| 165 |
246778_at |
AT5G27450
|
MK, mevalonate kinase, MVK, MEVALONATE KINASE |
-0.137 |
0.058 |
| 166 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.136 |
0.121 |
| 167 |
258697_at |
AT3G09660
|
AtMCM8, MCM8, minichromosome maintenance 8 |
-0.135 |
-0.019 |
| 168 |
250744_at |
AT5G05840
|
unknown |
-0.134 |
-0.282 |
| 169 |
260192_at |
AT1G67630
|
EMB2814, EMBRYO DEFECTIVE 2814, POLA2, DNA polymerase alpha 2 |
-0.133 |
0.096 |
| 170 |
259762_at |
AT1G77600
|
[ARM repeat superfamily protein] |
-0.132 |
0.013 |
| 171 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.132 |
0.108 |
| 172 |
249201_at |
AT5G42370
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.132 |
-0.010 |
| 173 |
247184_at |
AT5G65450
|
UBP17, ubiquitin-specific protease 17 |
-0.132 |
0.060 |
| 174 |
247503_at |
AT5G61980
|
AGD1, ARF-GAP domain 1 |
-0.131 |
0.037 |
| 175 |
258530_at |
AT3G06840
|
unknown |
-0.131 |
0.019 |
| 176 |
256429_at |
AT3G11040
|
AtENGase85B, ENGase85B, Endo-beta-N-acetyglucosaminidase 85B |
-0.130 |
-0.000 |
| 177 |
258424_at |
AT3G16750
|
unknown |
-0.130 |
0.007 |
| 178 |
253142_at |
AT4G35520
|
ATMLH3, MLH3, MUTL protein homolog 3 |
-0.128 |
0.099 |
| 179 |
256960_at |
AT3G13510
|
unknown |
-0.128 |
0.045 |
| 180 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.127 |
0.129 |
| 181 |
262619_at |
AT1G06550
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
-0.127 |
0.029 |
| 182 |
252100_at |
AT3G51110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.127 |
-0.069 |
| 183 |
252901_at |
AT4G39550
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.125 |
0.051 |
| 184 |
245789_at |
AT1G32090
|
[early-responsive to dehydration stress protein (ERD4)] |
-0.125 |
0.245 |
| 185 |
257848_at |
AT3G13030
|
[hAT transposon superfamily protein] |
-0.124 |
0.014 |
| 186 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.123 |
-0.114 |
| 187 |
246082_at |
AT5G20480
|
EFR, EF-TU receptor |
-0.122 |
-0.157 |
| 188 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.122 |
0.040 |
| 189 |
265897_at |
AT2G25680
|
MOT1, molybdate transporter 1 |
-0.122 |
-0.022 |
| 190 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.121 |
-0.112 |
| 191 |
256924_at |
AT3G29590
|
AT5MAT |
-0.121 |
0.100 |
| 192 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
-0.121 |
-0.095 |
| 193 |
259285_at |
AT3G11460
|
MEF10, mitochondrial RNA editing factor 10 |
-0.120 |
0.115 |
| 194 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.120 |
0.006 |
| 195 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.120 |
-0.124 |
| 196 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.119 |
0.019 |
| 197 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.119 |
0.246 |
| 198 |
251733_at |
AT3G56240
|
CCH, copper chaperone |
-0.119 |
-0.031 |
| 199 |
266002_at |
AT2G37310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.119 |
0.155 |
| 200 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-0.119 |
0.158 |
| 201 |
261972_at |
AT1G64600
|
[methyltransferases] |
-0.118 |
0.078 |
| 202 |
265277_at |
AT2G28410
|
unknown |
-0.117 |
0.060 |
| 203 |
262907_at |
AT1G59720
|
CRR28, CHLORORESPIRATORY REDUCTION28 |
-0.116 |
0.026 |
| 204 |
246188_at |
AT5G21050
|
[LOCATED IN: chloroplast] |
-0.115 |
0.001 |
| 205 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
-0.115 |
0.029 |
| 206 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.114 |
-0.108 |
| 207 |
265708_at |
AT2G03380
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.114 |
0.040 |
| 208 |
250720_at |
AT5G06180
|
unknown |
-0.114 |
-0.036 |
| 209 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
-0.114 |
0.003 |
| 210 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.113 |
-0.156 |
| 211 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
-0.113 |
-0.041 |
| 212 |
252194_at |
AT3G50110
|
ATPEN3, Arabidopsis thaliana phosphatase and TENsin homolog deleted on chromosome ten 3, PEN3, PTEN 3, PTEN2B, phosphatase and TENsin homolog deleted on chromosome ten 2B |
-0.113 |
0.002 |
| 213 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
-0.113 |
0.061 |