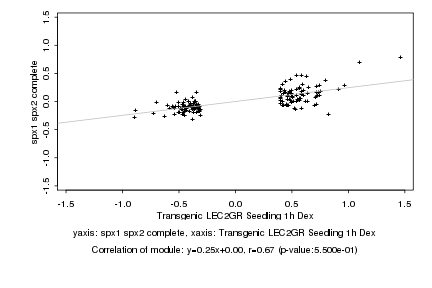
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Transgenic LEC2GR Seedling 1h Dex |
Log2 signal ratio
spx1 spx2 complete |
| 1 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
1.458 |
0.790 |
| 2 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
1.098 |
0.708 |
| 3 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.957 |
0.294 |
| 4 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.910 |
0.220 |
| 5 |
254574_at |
AT4G19430
|
unknown |
0.822 |
-0.229 |
| 6 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.791 |
0.382 |
| 7 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.748 |
0.190 |
| 8 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.742 |
0.291 |
| 9 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.736 |
0.118 |
| 10 |
245041_at |
AT2G26530
|
AR781 |
0.733 |
0.174 |
| 11 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.726 |
0.110 |
| 12 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.716 |
-0.051 |
| 13 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.712 |
0.270 |
| 14 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.709 |
0.097 |
| 15 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.709 |
0.167 |
| 16 |
262801_at |
AT1G21010
|
unknown |
0.701 |
0.079 |
| 17 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
0.697 |
-0.058 |
| 18 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.642 |
0.258 |
| 19 |
261662_at |
AT1G18350
|
ATMKK7, MAP kinase kinase 7, BUD1, BUSHY AND DWARF 1, MKK7, MAP kinase kinase 7, MKK7, MAP KINASE KINASE7 |
0.632 |
0.005 |
| 20 |
247933_at |
AT5G56980
|
unknown |
0.630 |
0.151 |
| 21 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.621 |
0.460 |
| 22 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.619 |
0.003 |
| 23 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.599 |
0.202 |
| 24 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.597 |
0.119 |
| 25 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.591 |
0.316 |
| 26 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.584 |
-0.116 |
| 27 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
0.579 |
0.182 |
| 28 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.579 |
0.133 |
| 29 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.578 |
0.476 |
| 30 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.577 |
0.153 |
| 31 |
253830_at |
AT4G27652
|
unknown |
0.569 |
0.069 |
| 32 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
0.566 |
0.019 |
| 33 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.566 |
0.238 |
| 34 |
248889_at |
AT5G46230
|
unknown |
0.562 |
0.257 |
| 35 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.560 |
0.071 |
| 36 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
0.549 |
0.037 |
| 37 |
252661_at |
AT3G44450
|
unknown |
0.540 |
0.468 |
| 38 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.538 |
0.231 |
| 39 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.537 |
0.115 |
| 40 |
248286_at |
AT5G52870
|
MAKR5, MEMBRANE-ASSOCIATED KINASE REGULATOR 5 |
0.537 |
0.016 |
| 41 |
265732_at |
AT2G01300
|
unknown |
0.531 |
-0.128 |
| 42 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.517 |
-0.116 |
| 43 |
260528_at |
AT2G47260
|
ATWRKY23, WRKY DNA-BINDING PROTEIN 23, WRKY23, WRKY DNA-binding protein 23 |
0.507 |
0.005 |
| 44 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.498 |
0.094 |
| 45 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.497 |
0.087 |
| 46 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.493 |
0.149 |
| 47 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
0.492 |
0.099 |
| 48 |
247177_at |
AT5G65300
|
unknown |
0.491 |
0.198 |
| 49 |
250098_at |
AT5G17350
|
unknown |
0.489 |
-0.008 |
| 50 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
0.489 |
-0.015 |
| 51 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
0.484 |
0.143 |
| 52 |
261247_at |
AT1G20070
|
unknown |
0.482 |
0.026 |
| 53 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.479 |
0.407 |
| 54 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.477 |
0.179 |
| 55 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.477 |
0.053 |
| 56 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
0.468 |
-0.070 |
| 57 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.464 |
0.150 |
| 58 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.457 |
0.100 |
| 59 |
266536_at |
AT2G16900
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.456 |
0.046 |
| 60 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.456 |
-0.060 |
| 61 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
0.454 |
0.120 |
| 62 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.451 |
-0.043 |
| 63 |
257480_at |
AT1G15640
|
unknown |
0.450 |
-0.070 |
| 64 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
0.440 |
0.177 |
| 65 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.440 |
0.360 |
| 66 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.427 |
0.204 |
| 67 |
248277_at |
AT5G52860
|
ABCG8, ATP-binding cassette G8 |
0.424 |
-0.056 |
| 68 |
245740_at |
AT1G44100
|
AAP5, amino acid permease 5 |
0.422 |
0.147 |
| 69 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.421 |
-0.060 |
| 70 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.416 |
0.008 |
| 71 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.412 |
0.305 |
| 72 |
245677_at |
AT1G56660
|
unknown |
0.410 |
-0.055 |
| 73 |
260387_at |
AT1G74100
|
ATSOT16, SULFOTRANSFERASE 16, ATST5A, ARABIDOPSIS SULFOTRANSFERASE 5A, CORI-7, CORONATINE INDUCED-7, SOT16, sulfotransferase 16 |
0.407 |
0.112 |
| 74 |
266882_at |
AT2G44670
|
unknown |
0.398 |
0.028 |
| 75 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.397 |
0.071 |
| 76 |
264535_at |
AT1G55690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.395 |
-0.029 |
| 77 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.395 |
0.225 |
| 78 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.393 |
0.248 |
| 79 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.392 |
0.181 |
| 80 |
251653_at |
AT3G57130
|
BOP1, BLADE ON PETIOLE 1 |
0.392 |
0.089 |
| 81 |
261567_at |
AT1G33055
|
unknown |
-0.902 |
-0.272 |
| 82 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.892 |
-0.149 |
| 83 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.732 |
-0.198 |
| 84 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.703 |
-0.006 |
| 85 |
259996_at |
AT1G67910
|
unknown |
-0.633 |
-0.250 |
| 86 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.604 |
-0.053 |
| 87 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.584 |
-0.109 |
| 88 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.558 |
-0.076 |
| 89 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.555 |
-0.093 |
| 90 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.547 |
-0.115 |
| 91 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.545 |
-0.227 |
| 92 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.539 |
-0.088 |
| 93 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.528 |
0.168 |
| 94 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.512 |
-0.078 |
| 95 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
-0.511 |
-0.012 |
| 96 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.506 |
-0.182 |
| 97 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.497 |
-0.190 |
| 98 |
259235_at |
AT3G11600
|
unknown |
-0.492 |
-0.110 |
| 99 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.481 |
-0.134 |
| 100 |
266259_at |
AT2G27830
|
unknown |
-0.471 |
-0.052 |
| 101 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.469 |
-0.227 |
| 102 |
253246_at |
AT4G34600
|
unknown |
-0.469 |
-0.136 |
| 103 |
266635_at |
AT2G35470
|
unknown |
-0.468 |
-0.080 |
| 104 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.460 |
-0.182 |
| 105 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.460 |
-0.007 |
| 106 |
257946_at |
AT3G21710
|
unknown |
-0.459 |
-0.174 |
| 107 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.457 |
-0.239 |
| 108 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.455 |
-0.241 |
| 109 |
254193_at |
AT4G23870
|
unknown |
-0.453 |
-0.082 |
| 110 |
248969_at |
AT5G45310
|
unknown |
-0.452 |
-0.135 |
| 111 |
263265_at |
AT2G38820
|
unknown |
-0.452 |
-0.103 |
| 112 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.451 |
-0.072 |
| 113 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
-0.449 |
0.046 |
| 114 |
250008_at |
AT5G18630
|
[alpha/beta-Hydrolases superfamily protein] |
-0.448 |
-0.093 |
| 115 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.440 |
-0.157 |
| 116 |
250189_at |
AT5G14410
|
unknown |
-0.435 |
-0.113 |
| 117 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.421 |
-0.082 |
| 118 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.419 |
0.016 |
| 119 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.415 |
-0.055 |
| 120 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.410 |
-0.165 |
| 121 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.404 |
-0.055 |
| 122 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.402 |
-0.025 |
| 123 |
259541_at |
AT1G20650
|
ASG5, ALTERED SEED GERMINATION 5 |
-0.397 |
-0.084 |
| 124 |
263333_at |
AT2G03890
|
ATPI4K GAMMA 7, phosphoinositide 4-kinase gamma 7, PI4K GAMMA 7, phosphoinositide 4-kinase gamma 7, UBDK GAMMA 7, UBIQUITIN-LIKE DOMAIN KINASE GAMMA 7 |
-0.395 |
-0.080 |
| 125 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.393 |
-0.096 |
| 126 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.389 |
-0.315 |
| 127 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.388 |
-0.092 |
| 128 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.387 |
0.086 |
| 129 |
246244_at |
AT4G37250
|
[Leucine-rich repeat protein kinase family protein] |
-0.384 |
-0.140 |
| 130 |
248795_at |
AT5G47390
|
MYBH, MYB hypocotyl elongation-related |
-0.382 |
-0.087 |
| 131 |
259017_at |
AT3G07310
|
unknown |
-0.377 |
-0.114 |
| 132 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.375 |
-0.030 |
| 133 |
245984_at |
AT5G13090
|
unknown |
-0.375 |
-0.104 |
| 134 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.374 |
-0.194 |
| 135 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.373 |
-0.088 |
| 136 |
246917_at |
AT5G25280
|
[serine-rich protein-related] |
-0.369 |
-0.108 |
| 137 |
264040_at |
AT2G03730
|
ACR5, ACT domain repeat 5 |
-0.366 |
-0.137 |
| 138 |
245602_at |
AT4G14270
|
unknown |
-0.364 |
0.001 |
| 139 |
262286_at |
AT1G68585
|
unknown |
-0.361 |
0.004 |
| 140 |
267591_at |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-0.357 |
0.010 |
| 141 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.352 |
-0.092 |
| 142 |
258225_at |
AT3G15630
|
unknown |
-0.352 |
-0.039 |
| 143 |
246975_at |
AT5G24890
|
unknown |
-0.351 |
-0.060 |
| 144 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.350 |
-0.052 |
| 145 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
-0.346 |
0.168 |
| 146 |
263118_at |
AT1G03090
|
MCCA |
-0.345 |
-0.178 |
| 147 |
265457_at |
AT2G46550
|
unknown |
-0.344 |
-0.065 |
| 148 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.336 |
-0.097 |
| 149 |
257990_at |
AT3G19860
|
bHLH121, basic Helix-Loop-Helix 121 |
-0.334 |
-0.172 |
| 150 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.331 |
-0.048 |
| 151 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
-0.328 |
-0.099 |
| 152 |
263827_at |
AT2G40420
|
[Transmembrane amino acid transporter family protein] |
-0.327 |
-0.133 |
| 153 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.326 |
-0.140 |
| 154 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.323 |
-0.131 |
| 155 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.323 |
-0.143 |
| 156 |
258350_at |
AT3G17510
|
CIPK1, CBL-interacting protein kinase 1, SnRK3.16, SNF1-RELATED PROTEIN KINASE 3.16 |
-0.322 |
-0.164 |
| 157 |
263799_at |
AT2G24550
|
unknown |
-0.322 |
-0.076 |
| 158 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
-0.317 |
-0.128 |
| 159 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.314 |
-0.248 |