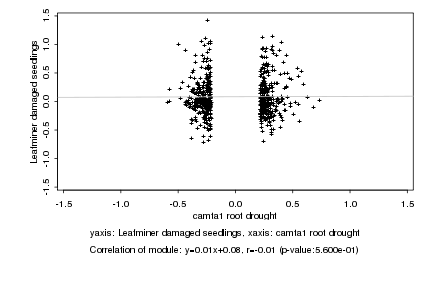
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
camta1 root drought |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.732 |
0.020 |
| 2 |
258383_at |
AT3G15440
|
unknown |
0.678 |
-0.089 |
| 3 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.626 |
0.075 |
| 4 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.586 |
0.307 |
| 5 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.571 |
0.535 |
| 6 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.558 |
-0.339 |
| 7 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.550 |
0.441 |
| 8 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
0.544 |
-0.052 |
| 9 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.534 |
0.582 |
| 10 |
250940_at |
AT5G03310
|
SAUR44, SMALL AUXIN UPREGULATED RNA 44 |
0.517 |
-0.017 |
| 11 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.501 |
-0.211 |
| 12 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.482 |
0.402 |
| 13 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
0.476 |
-0.077 |
| 14 |
247320_at |
AT5G64040
|
PSAN |
0.472 |
-0.028 |
| 15 |
263881_at |
AT2G21820
|
unknown |
0.465 |
0.408 |
| 16 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.453 |
0.500 |
| 17 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.450 |
0.072 |
| 18 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.442 |
0.261 |
| 19 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.441 |
0.274 |
| 20 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.439 |
0.823 |
| 21 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.429 |
-0.149 |
| 22 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.428 |
0.498 |
| 23 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.426 |
0.236 |
| 24 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.425 |
-0.142 |
| 25 |
259552_at |
AT1G21320
|
[nucleotide binding] |
0.425 |
-0.038 |
| 26 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.411 |
0.701 |
| 27 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.408 |
0.060 |
| 28 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.407 |
-0.319 |
| 29 |
258631_at |
AT3G07970
|
QRT2, QUARTET 2 |
0.404 |
0.046 |
| 30 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.401 |
1.046 |
| 31 |
261817_at |
AT1G08180
|
unknown |
0.399 |
-0.271 |
| 32 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
0.397 |
0.090 |
| 33 |
263281_at |
AT2G14160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.395 |
0.002 |
| 34 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.389 |
0.487 |
| 35 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.389 |
-0.244 |
| 36 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
0.386 |
-0.442 |
| 37 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.384 |
0.903 |
| 38 |
257172_at |
AT3G23700
|
[Nucleic acid-binding proteins superfamily] |
0.382 |
-0.205 |
| 39 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
0.376 |
-0.004 |
| 40 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.374 |
-0.020 |
| 41 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.369 |
0.043 |
| 42 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
0.364 |
0.324 |
| 43 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
0.362 |
0.068 |
| 44 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
0.361 |
0.042 |
| 45 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.358 |
0.318 |
| 46 |
250624_at |
AT5G07330
|
unknown |
0.355 |
0.011 |
| 47 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
0.355 |
-0.080 |
| 48 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.353 |
0.811 |
| 49 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.353 |
-0.152 |
| 50 |
253823_at |
AT4G28030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.349 |
-0.125 |
| 51 |
259896_at |
AT1G71500
|
[Rieske (2Fe-2S) domain-containing protein] |
0.343 |
-0.163 |
| 52 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
0.337 |
-0.235 |
| 53 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
0.334 |
0.328 |
| 54 |
248205_at |
AT5G54300
|
unknown |
0.334 |
0.559 |
| 55 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.332 |
-0.068 |
| 56 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
0.329 |
-0.321 |
| 57 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.327 |
0.856 |
| 58 |
263736_at |
AT1G60000
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.327 |
-0.260 |
| 59 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.325 |
-0.481 |
| 60 |
245330_at |
AT4G14930
|
[Survival protein SurE-like phosphatase/nucleotidase] |
0.324 |
-0.028 |
| 61 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.324 |
0.050 |
| 62 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.323 |
0.259 |
| 63 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.322 |
1.152 |
| 64 |
252036_at |
AT3G52070
|
unknown |
0.321 |
0.095 |
| 65 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.319 |
0.968 |
| 66 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
0.319 |
-0.168 |
| 67 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.318 |
0.182 |
| 68 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.318 |
0.902 |
| 69 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.317 |
0.559 |
| 70 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.315 |
0.622 |
| 71 |
260635_at |
AT1G62422
|
unknown |
0.313 |
0.042 |
| 72 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
0.312 |
-0.486 |
| 73 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.312 |
-0.561 |
| 74 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.310 |
0.324 |
| 75 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.310 |
-0.016 |
| 76 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.310 |
0.916 |
| 77 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.309 |
-0.206 |
| 78 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
0.308 |
0.256 |
| 79 |
256222_at |
AT1G56210
|
[Heavy metal transport/detoxification superfamily protein ] |
0.308 |
-0.532 |
| 80 |
267003_at |
AT2G34340
|
unknown |
0.307 |
-0.193 |
| 81 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
0.307 |
-0.277 |
| 82 |
251727_at |
AT3G56290
|
unknown |
0.307 |
-0.250 |
| 83 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.306 |
0.285 |
| 84 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.304 |
-0.416 |
| 85 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
0.303 |
-0.114 |
| 86 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.301 |
-0.184 |
| 87 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
0.300 |
-0.245 |
| 88 |
246054_at |
AT5G08360
|
unknown |
0.300 |
-0.042 |
| 89 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.299 |
-0.217 |
| 90 |
263147_at |
AT1G53980
|
[Ubiquitin-like superfamily protein] |
0.298 |
0.041 |
| 91 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
0.296 |
-0.297 |
| 92 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.296 |
0.080 |
| 93 |
254020_at |
AT4G25700
|
B1, BCH1, BETA CAROTENOID HYDROXYLASE 1, BETA-OHASE 1, beta-hydroxylase 1, chy1 |
0.295 |
-0.359 |
| 94 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.293 |
-0.340 |
| 95 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.292 |
0.288 |
| 96 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.289 |
0.245 |
| 97 |
265983_at |
AT2G18550
|
ATHB21, homeobox protein 21, HB-2, homeobox-2, HB21, homeobox protein 21 |
0.288 |
0.018 |
| 98 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
0.287 |
0.091 |
| 99 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.286 |
0.067 |
| 100 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
0.286 |
-0.263 |
| 101 |
259193_at |
AT3G01480
|
ATCYP38, ARABIDOPSIS CYCLOPHILIN 38, CYP38, cyclophilin 38 |
0.286 |
-0.174 |
| 102 |
258993_at |
AT3G08940
|
LHCB4.2, light harvesting complex photosystem II |
0.286 |
-0.064 |
| 103 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.284 |
0.653 |
| 104 |
257832_at |
AT3G26740
|
CCL, CCR-like |
0.283 |
0.512 |
| 105 |
250421_at |
AT5G11270
|
OCP3, overexpressor of cationic peroxidase 3 |
0.282 |
-0.215 |
| 106 |
256097_at |
AT1G13670
|
unknown |
0.282 |
-0.005 |
| 107 |
251194_at |
AT3G62920
|
unknown |
0.282 |
0.067 |
| 108 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.281 |
-0.032 |
| 109 |
249941_at |
AT5G22270
|
unknown |
0.281 |
0.470 |
| 110 |
266551_at |
AT2G35260
|
unknown |
0.281 |
-0.155 |
| 111 |
260389_at |
AT1G74055
|
unknown |
0.280 |
-0.080 |
| 112 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
0.278 |
-0.018 |
| 113 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.277 |
0.623 |
| 114 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.277 |
-0.157 |
| 115 |
257205_at |
AT3G16520
|
UGT88A1, UDP-glucosyl transferase 88A1 |
0.276 |
-0.086 |
| 116 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.276 |
0.161 |
| 117 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.275 |
0.558 |
| 118 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
0.275 |
-0.271 |
| 119 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
0.273 |
0.027 |
| 120 |
261266_at |
AT1G26770
|
AT-EXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXP10, ARABIDOPSIS THALIANA EXPANSIN 10, ATEXPA10, expansin A10, ATHEXP ALPHA 1.1, ARABIDOPSIS THALIANA EXPANSIN ALPHA 1.1, EXP10, EXPANSIN 10, EXPA10, expansin A10 |
0.273 |
0.219 |
| 121 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.273 |
0.101 |
| 122 |
259982_at |
AT1G76410
|
ATL8 |
0.273 |
0.077 |
| 123 |
249415_at |
AT5G39660
|
CDF2, cycling DOF factor 2 |
0.273 |
-0.414 |
| 124 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
0.272 |
0.207 |
| 125 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.272 |
0.192 |
| 126 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.272 |
0.104 |
| 127 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
0.271 |
-0.042 |
| 128 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.269 |
-0.206 |
| 129 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.269 |
0.097 |
| 130 |
258571_at |
AT3G04420
|
anac048, NAC domain containing protein 48, NAC048, NAC domain containing protein 48 |
0.269 |
-0.045 |
| 131 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.269 |
0.207 |
| 132 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.269 |
0.942 |
| 133 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
0.268 |
-0.220 |
| 134 |
257567_at |
AT3G23930
|
unknown |
0.268 |
-0.143 |
| 135 |
260841_at |
AT1G29195
|
unknown |
0.268 |
0.081 |
| 136 |
262879_at |
AT1G64860
|
RPOD1, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, sigma factor A, SIGB |
0.268 |
-0.204 |
| 137 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.268 |
0.081 |
| 138 |
252904_at |
AT4G39620
|
ATPPR5, A. THALIANA PENTATRICOPEPTIDE REPEAT 5, EMB2453, EMBRYO DEFECTIVE 2453 |
0.268 |
-0.306 |
| 139 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
0.268 |
0.260 |
| 140 |
254691_at |
AT4G17840
|
unknown |
0.267 |
-0.080 |
| 141 |
250951_at |
AT5G03550
|
unknown |
0.266 |
-0.079 |
| 142 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.266 |
0.515 |
| 143 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.266 |
0.783 |
| 144 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.266 |
0.668 |
| 145 |
265023_at |
AT1G24440
|
[RING/U-box superfamily protein] |
0.266 |
0.101 |
| 146 |
246929_at |
AT5G25210
|
unknown |
0.265 |
0.213 |
| 147 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
0.265 |
-0.036 |
| 148 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
0.263 |
-0.030 |
| 149 |
265431_at |
AT2G20680
|
AtMAN2, MAN2, endo-beta-mannase 2 |
0.262 |
-0.012 |
| 150 |
260688_at |
AT1G17665
|
unknown |
0.262 |
0.022 |
| 151 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.262 |
-0.309 |
| 152 |
256205_at |
AT1G50890
|
[ARM repeat superfamily protein] |
0.261 |
-0.287 |
| 153 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.260 |
0.894 |
| 154 |
259491_at |
AT1G15820
|
CP24, LHCB6, light harvesting complex photosystem II subunit 6 |
0.259 |
-0.051 |
| 155 |
259889_at |
AT1G76405
|
unknown |
0.259 |
-0.183 |
| 156 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.258 |
-0.284 |
| 157 |
260536_at |
AT2G43400
|
ETFQO, electron-transfer flavoprotein:ubiquinone oxidoreductase |
0.257 |
-0.076 |
| 158 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
0.256 |
-0.327 |
| 159 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.255 |
0.190 |
| 160 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.255 |
-0.327 |
| 161 |
250937_at |
AT5G03230
|
unknown |
0.255 |
0.322 |
| 162 |
249750_at |
AT5G24570
|
unknown |
0.255 |
-0.191 |
| 163 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.255 |
0.643 |
| 164 |
258398_at |
AT3G15360
|
ATHM4, ATM4, ARABIDOPSIS THIOREDOXIN M-TYPE 4, TRX-M4, thioredoxin M-type 4 |
0.254 |
0.025 |
| 165 |
267084_at |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
0.253 |
-0.099 |
| 166 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.253 |
0.322 |
| 167 |
260522_x_at |
AT2G41730
|
unknown |
0.253 |
0.082 |
| 168 |
254756_at |
AT4G13235
|
EDA21, embryo sac development arrest 21 |
0.252 |
0.024 |
| 169 |
251012_at |
AT5G02580
|
unknown |
0.252 |
0.353 |
| 170 |
259913_at |
AT1G72660
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.251 |
-0.105 |
| 171 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.251 |
0.220 |
| 172 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
0.250 |
0.039 |
| 173 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.249 |
0.156 |
| 174 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.249 |
-0.083 |
| 175 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.248 |
0.595 |
| 176 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.248 |
0.280 |
| 177 |
257670_at |
AT3G20340
|
unknown |
0.247 |
0.311 |
| 178 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.246 |
0.417 |
| 179 |
247283_at |
AT5G64250
|
[Aldolase-type TIM barrel family protein] |
0.246 |
0.105 |
| 180 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.246 |
0.527 |
| 181 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
0.246 |
-0.090 |
| 182 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.246 |
0.291 |
| 183 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.246 |
0.240 |
| 184 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.246 |
-0.125 |
| 185 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.246 |
0.773 |
| 186 |
249379_at |
AT5G40460
|
unknown |
0.245 |
0.416 |
| 187 |
263827_at |
AT2G40420
|
[Transmembrane amino acid transporter family protein] |
0.245 |
0.049 |
| 188 |
250563_at |
AT5G08050
|
unknown |
0.244 |
-0.160 |
| 189 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
0.243 |
-0.204 |
| 190 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.243 |
-0.276 |
| 191 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
0.242 |
0.058 |
| 192 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
0.242 |
-0.158 |
| 193 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.242 |
0.653 |
| 194 |
247754_at |
AT5G59080
|
unknown |
0.242 |
0.178 |
| 195 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.241 |
-0.272 |
| 196 |
253164_at |
AT4G35725
|
unknown |
0.241 |
-0.025 |
| 197 |
266016_at |
AT2G18670
|
[RING/U-box superfamily protein] |
0.240 |
0.429 |
| 198 |
261667_at |
AT1G18460
|
[alpha/beta-Hydrolases superfamily protein] |
0.239 |
-0.174 |
| 199 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.239 |
-0.088 |
| 200 |
245396_at |
AT4G14870
|
SECE1 |
0.238 |
-0.148 |
| 201 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.238 |
-0.049 |
| 202 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.238 |
-0.687 |
| 203 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.238 |
0.384 |
| 204 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.238 |
0.501 |
| 205 |
263460_at |
AT2G31810
|
[ACT domain-containing small subunit of acetolactate synthase protein] |
0.238 |
0.036 |
| 206 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
0.238 |
-0.000 |
| 207 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.237 |
0.300 |
| 208 |
264779_at |
AT1G08570
|
ACHT4, atypical CYS HIS rich thioredoxin 4 |
0.237 |
-0.260 |
| 209 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.237 |
0.038 |
| 210 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.237 |
0.102 |
| 211 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.237 |
0.935 |
| 212 |
260036_at |
AT1G68830
|
STN7, STT7 homolog STN7 |
0.237 |
-0.108 |
| 213 |
248215_at |
AT5G53680
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.237 |
0.035 |
| 214 |
266100_at |
AT2G37980
|
[O-fucosyltransferase family protein] |
0.236 |
0.344 |
| 215 |
250223_at |
AT5G14070
|
ROXY2 |
0.236 |
-0.033 |
| 216 |
247650_at |
AT5G59960
|
unknown |
0.235 |
-0.080 |
| 217 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
0.235 |
0.113 |
| 218 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.235 |
0.270 |
| 219 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.234 |
0.003 |
| 220 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.234 |
0.786 |
| 221 |
258094_at |
AT3G14690
|
CYP72A15, cytochrome P450, family 72, subfamily A, polypeptide 15 |
0.233 |
-0.079 |
| 222 |
263020_at |
AT1G23880
|
[NHL domain-containing protein] |
0.233 |
-0.182 |
| 223 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.232 |
0.609 |
| 224 |
254187_at |
AT4G23890
|
CRR31, CHLORORESPIRATORY REDUCTION 31, NdhS, NADH dehydrogenase-like complex S |
0.232 |
-0.143 |
| 225 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.232 |
0.039 |
| 226 |
263647_at |
AT2G04690
|
Pyridoxamine 5'-phosphate oxidase family protein |
0.232 |
0.010 |
| 227 |
245000_at |
ATCG00210
|
YCF6 |
0.231 |
-0.087 |
| 228 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
0.231 |
-0.045 |
| 229 |
263985_at |
AT2G42750
|
DJC77, DNA J protein C77 |
0.231 |
-0.194 |
| 230 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.230 |
-0.517 |
| 231 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
0.230 |
0.172 |
| 232 |
264828_at |
AT1G03380
|
ATATG18G, homolog of yeast autophagy 18 (ATG18) G, ATG18G, homolog of yeast autophagy 18 (ATG18) G |
0.230 |
-0.127 |
| 233 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.230 |
0.595 |
| 234 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
0.230 |
-0.174 |
| 235 |
261086_at |
AT1G17460
|
TRFL3, TRF-like 3 |
0.230 |
-0.120 |
| 236 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.229 |
0.043 |
| 237 |
262638_at |
AT1G06650
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.229 |
0.081 |
| 238 |
250766_at |
AT5G05550
|
[sequence-specific DNA binding transcription factors] |
0.229 |
-0.099 |
| 239 |
267311_at |
AT2G34670
|
unknown |
0.229 |
-0.014 |
| 240 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.229 |
1.135 |
| 241 |
244932_at |
ATCG01060
|
PSAC |
0.228 |
-0.063 |
| 242 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.228 |
0.931 |
| 243 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
0.228 |
0.180 |
| 244 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.228 |
-0.265 |
| 245 |
245388_at |
AT4G16410
|
unknown |
0.228 |
-0.122 |
| 246 |
267300_at |
AT2G30140
|
UGT87A2, UDP-glucosyl transferase 87A2 |
0.228 |
0.221 |
| 247 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.228 |
0.928 |
| 248 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.228 |
0.312 |
| 249 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.227 |
-0.053 |
| 250 |
248154_at |
AT5G54400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.227 |
0.026 |
| 251 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.227 |
-0.256 |
| 252 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
0.227 |
-0.438 |
| 253 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.226 |
-0.149 |
| 254 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.225 |
0.024 |
| 255 |
247783_at |
AT5G58800
|
[Quinone reductase family protein] |
0.225 |
-0.052 |
| 256 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
0.225 |
-0.381 |
| 257 |
256765_at |
AT3G22200
|
GABA-T, GAMMA-AMINOBUTYRATE TRANSAMINASE, HER1, HEXENAL RESPONSE1, POP2, POLLEN-PISTIL INCOMPATIBILITY 2 |
0.224 |
0.173 |
| 258 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.224 |
-0.333 |
| 259 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.223 |
0.792 |
| 260 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
0.223 |
-0.029 |
| 261 |
245487_at |
AT4G16250
|
PHYD, phytochrome D |
0.223 |
0.029 |
| 262 |
246524_at |
AT5G15860
|
ATPCME, prenylcysteine methylesterase, ICME, Isoprenylcysteine methylesterase, PCME, prenylcysteine methylesterase |
0.223 |
-0.263 |
| 263 |
259637_at |
AT1G52260
|
ATPDI3, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 3, ATPDIL1-5, PDI-like 1-5, PDI3, PROTEIN DISULFIDE ISOMERASE 3, PDIL1-5, PDI-like 1-5 |
0.222 |
-0.069 |
| 264 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
0.222 |
-0.158 |
| 265 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
0.222 |
0.007 |
| 266 |
261319_at |
AT1G53090
|
SPA4, SPA1-related 4 |
0.222 |
-0.215 |
| 267 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.222 |
-0.163 |
| 268 |
259421_at |
AT1G13910
|
[Leucine-rich repeat (LRR) family protein] |
0.221 |
-0.148 |
| 269 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.221 |
-0.261 |
| 270 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.221 |
0.563 |
| 271 |
259738_at |
AT1G64355
|
unknown |
0.220 |
-0.170 |
| 272 |
246278_at |
AT4G37190
|
[LOCATED IN: cytosol, plasma membrane] |
0.220 |
0.304 |
| 273 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.219 |
0.212 |
| 274 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.219 |
0.488 |
| 275 |
256069_at |
AT1G13740
|
AFP2, ABI five binding protein 2 |
0.219 |
0.292 |
| 276 |
258110_at |
AT3G14610
|
CYP72A7, cytochrome P450, family 72, subfamily A, polypeptide 7 |
0.219 |
-0.006 |
| 277 |
263118_at |
AT1G03090
|
MCCA |
0.218 |
0.199 |
| 278 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.218 |
0.154 |
| 279 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.218 |
0.543 |
| 280 |
251719_at |
AT3G56140
|
unknown |
0.218 |
-0.203 |
| 281 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.218 |
0.150 |
| 282 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
0.218 |
-0.195 |
| 283 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
0.218 |
-0.212 |
| 284 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.217 |
0.063 |
| 285 |
260044_at |
AT1G73655
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.215 |
-0.135 |
| 286 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.215 |
-0.217 |
| 287 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.215 |
0.011 |
| 288 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.215 |
-0.253 |
| 289 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.215 |
-0.224 |
| 290 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.215 |
0.082 |
| 291 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
0.215 |
-0.128 |
| 292 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
0.214 |
-0.058 |
| 293 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.214 |
0.452 |
| 294 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
0.213 |
-0.039 |
| 295 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
0.213 |
-0.208 |
| 296 |
248167_at |
AT5G54530
|
unknown |
0.213 |
-0.315 |
| 297 |
254178_at |
AT4G23880
|
unknown |
0.213 |
0.289 |
| 298 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
0.213 |
0.234 |
| 299 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.212 |
0.015 |
| 300 |
260898_at |
AT1G29070
|
[Ribosomal protein L34] |
0.211 |
-0.123 |
| 301 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.596 |
-0.009 |
| 302 |
264282_at |
AT1G61840
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.584 |
0.016 |
| 303 |
249522_at |
AT5G38700
|
unknown |
-0.576 |
0.219 |
| 304 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.498 |
1.005 |
| 305 |
266011_at |
AT2G37440
|
[DNAse I-like superfamily protein] |
-0.484 |
0.067 |
| 306 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-0.480 |
0.238 |
| 307 |
252084_at |
AT3G51970
|
ASAT1, acyl-CoA sterol acyl transferase 1, ATASAT1, ATSAT1, ARABIDOPSIS THALIANA STEROL O-ACYLTRANSFERASE 1 |
-0.463 |
0.342 |
| 308 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.441 |
-0.012 |
| 309 |
266455_at |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.440 |
0.899 |
| 310 |
264495_at |
AT1G27380
|
RIC2, ROP-interactive CRIB motif-containing protein 2 |
-0.436 |
-0.038 |
| 311 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.433 |
-0.054 |
| 312 |
266222_at |
AT2G28780
|
unknown |
-0.432 |
0.005 |
| 313 |
261575_at |
AT1G01130
|
unknown |
-0.418 |
-0.012 |
| 314 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.417 |
-0.023 |
| 315 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.414 |
0.265 |
| 316 |
253937_at |
AT4G26890
|
MAPKKK16, mitogen-activated protein kinase kinase kinase 16 |
-0.412 |
0.033 |
| 317 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
-0.408 |
-0.091 |
| 318 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
-0.402 |
0.088 |
| 319 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
-0.397 |
0.431 |
| 320 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.397 |
-0.039 |
| 321 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.395 |
-0.087 |
| 322 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.394 |
0.421 |
| 323 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
-0.389 |
0.187 |
| 324 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.389 |
-0.311 |
| 325 |
249897_at |
AT5G22550
|
unknown |
-0.387 |
0.058 |
| 326 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.385 |
-0.381 |
| 327 |
246001_at |
AT5G20790
|
unknown |
-0.385 |
-0.646 |
| 328 |
256703_at |
AT3G30260
|
AGL79, AGAMOUS-like 79 |
-0.381 |
-0.013 |
| 329 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.379 |
-0.109 |
| 330 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.379 |
0.520 |
| 331 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.376 |
0.226 |
| 332 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.376 |
-0.327 |
| 333 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.372 |
0.555 |
| 334 |
253883_at |
AT4G27660
|
unknown |
-0.371 |
0.170 |
| 335 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.369 |
0.216 |
| 336 |
248307_at |
AT5G52850
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.364 |
-0.075 |
| 337 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.363 |
0.155 |
| 338 |
258119_at |
AT3G14720
|
ATMPK19, ARABIDOPSIS THALIANA MAP KINASE 19, MPK19, MAP kinase 19 |
-0.363 |
-0.013 |
| 339 |
248319_at |
AT5G52710
|
[Copper transport protein family] |
-0.360 |
-0.061 |
| 340 |
251207_at |
AT3G63050
|
unknown |
-0.359 |
0.224 |
| 341 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.359 |
-0.138 |
| 342 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.358 |
0.413 |
| 343 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.357 |
-0.142 |
| 344 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.356 |
-0.270 |
| 345 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.354 |
0.157 |
| 346 |
249246_at |
AT5G42290
|
[transcription activator-related] |
-0.354 |
-0.007 |
| 347 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
-0.353 |
-0.099 |
| 348 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.350 |
0.037 |
| 349 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.350 |
0.807 |
| 350 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.349 |
-0.311 |
| 351 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.349 |
-0.148 |
| 352 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.349 |
0.699 |
| 353 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.347 |
-0.061 |
| 354 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.346 |
0.020 |
| 355 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.345 |
-0.016 |
| 356 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.345 |
-0.075 |
| 357 |
261215_at |
AT1G32970
|
[Subtilisin-like serine endopeptidase family protein] |
-0.343 |
0.037 |
| 358 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.341 |
-0.049 |
| 359 |
254374_at |
AT4G21780
|
unknown |
-0.341 |
0.149 |
| 360 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.341 |
0.174 |
| 361 |
246520_at |
AT5G15790
|
[RING/U-box superfamily protein] |
-0.341 |
-0.349 |
| 362 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.339 |
-0.467 |
| 363 |
245034_at |
AT2G26390
|
[Serine protease inhibitor (SERPIN) family protein] |
-0.339 |
0.335 |
| 364 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.338 |
-0.065 |
| 365 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
-0.337 |
0.307 |
| 366 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.336 |
-0.100 |
| 367 |
253821_at |
AT4G28380
|
[Leucine-rich repeat (LRR) family protein] |
-0.335 |
-0.001 |
| 368 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.335 |
0.036 |
| 369 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.334 |
0.217 |
| 370 |
261703_at |
AT1G32770
|
ANAC012, NAC domain containing protein 12, NAC012, NAC domain containing protein 12, NST3, NAC SECONDARY WALL THICKENING PROMOTING 3, SND1, SECONDARY WALL-ASSOCIATED NAC DOMAIN 1 |
-0.333 |
0.027 |
| 371 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.332 |
0.047 |
| 372 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
-0.332 |
0.319 |
| 373 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.331 |
0.248 |
| 374 |
249094_at |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
-0.331 |
-0.015 |
| 375 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.330 |
-0.193 |
| 376 |
247020_at |
AT5G67020
|
unknown |
-0.327 |
0.156 |
| 377 |
261928_at |
AT1G22480
|
[Cupredoxin superfamily protein] |
-0.326 |
0.112 |
| 378 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.325 |
-0.001 |
| 379 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.322 |
0.248 |
| 380 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
-0.318 |
-0.188 |
| 381 |
252211_at |
AT3G50220
|
IRX15, IRREGULAR XYLEM 15 |
-0.317 |
-0.086 |
| 382 |
259260_at |
AT3G11370
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.317 |
0.008 |
| 383 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.316 |
0.013 |
| 384 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
-0.316 |
0.069 |
| 385 |
258551_at |
AT3G06890
|
unknown |
-0.316 |
0.159 |
| 386 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.314 |
0.286 |
| 387 |
263158_at |
AT1G54160
|
NF-YA5, nuclear factor Y, subunit A5, NFYA5, NUCLEAR FACTOR Y A5 |
-0.314 |
0.184 |
| 388 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.313 |
0.221 |
| 389 |
252070_at |
AT3G51680
|
AtSDR2, SDR2, short-chain dehydrogenase/reductase 2 |
-0.313 |
-0.008 |
| 390 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.311 |
-0.203 |
| 391 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.310 |
-0.411 |
| 392 |
257233_at |
AT3G15050
|
IQD10, IQ-domain 10 |
-0.310 |
-0.009 |
| 393 |
245728_at |
AT1G73340
|
[Cytochrome P450 superfamily protein] |
-0.308 |
0.001 |
| 394 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
-0.307 |
-0.023 |
| 395 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.307 |
0.021 |
| 396 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.306 |
0.415 |
| 397 |
249167_at |
AT5G42860
|
unknown |
-0.306 |
0.083 |
| 398 |
245910_at |
AT5G09410
|
CAMTA1, calmodulin-binding transcription activator 1, EICBP.B, ethylene induced calmodulin binding protein |
-0.305 |
-0.071 |
| 399 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.305 |
0.041 |
| 400 |
256763_at |
AT3G16860
|
COBL8, COBRA-like protein 8 precursor |
-0.304 |
0.598 |
| 401 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.304 |
0.000 |
| 402 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.302 |
0.035 |
| 403 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.302 |
0.141 |
| 404 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.302 |
-0.090 |
| 405 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
-0.301 |
1.068 |
| 406 |
261101_at |
AT1G63030
|
ddf2, DWARF AND DELAYED FLOWERING 2 |
-0.300 |
0.038 |
| 407 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.296 |
-0.017 |
| 408 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.295 |
0.155 |
| 409 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
-0.294 |
0.812 |
| 410 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-0.294 |
-0.045 |
| 411 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.293 |
0.030 |
| 412 |
262575_at |
AT1G15210
|
ABCG35, ATP-binding cassette G35, ATPDR7, PLEIOTROPIC DRUG RESISTANCE 7, PDR7, pleiotropic drug resistance 7 |
-0.292 |
0.076 |
| 413 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.292 |
-0.110 |
| 414 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
-0.291 |
0.019 |
| 415 |
253965_at |
AT4G26490
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.290 |
-0.158 |
| 416 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
-0.290 |
0.255 |
| 417 |
251925_at |
AT3G54000
|
unknown |
-0.289 |
0.060 |
| 418 |
247830_at |
AT5G58530
|
[Glutaredoxin family protein] |
-0.288 |
0.030 |
| 419 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
-0.288 |
-0.044 |
| 420 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.287 |
0.716 |
| 421 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.286 |
0.075 |
| 422 |
259850_at |
AT1G72240
|
unknown |
-0.286 |
0.165 |
| 423 |
247290_at |
AT5G64450
|
unknown |
-0.285 |
0.281 |
| 424 |
261278_at |
AT1G05800
|
DGL, DONGLE |
-0.284 |
0.339 |
| 425 |
245953_at |
AT5G28520
|
[Mannose-binding lectin superfamily protein] |
-0.283 |
0.107 |
| 426 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.282 |
0.176 |
| 427 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.280 |
0.067 |
| 428 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.280 |
-0.604 |
| 429 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
-0.280 |
0.718 |
| 430 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.280 |
-0.164 |
| 431 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.279 |
-0.713 |
| 432 |
258468_at |
AT3G06070
|
unknown |
-0.279 |
-0.278 |
| 433 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.279 |
0.250 |
| 434 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.279 |
0.068 |
| 435 |
267206_at |
AT2G30830
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.278 |
0.998 |
| 436 |
267555_at |
AT2G32765
|
ATSUMO5, SUM5, SUMO 5, SUMO5, small ubiquitinrelated modifier 5 |
-0.278 |
-0.287 |
| 437 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-0.278 |
-0.070 |
| 438 |
248741_at |
AT5G48170
|
SLY2, SLEEPY2, SNE, SNEEZY |
-0.278 |
-0.028 |
| 439 |
263545_at |
AT2G21560
|
unknown |
-0.278 |
-0.365 |
| 440 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.276 |
-0.440 |
| 441 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-0.276 |
0.014 |
| 442 |
251610_at |
AT3G57930
|
unknown |
-0.276 |
0.017 |
| 443 |
262625_at |
AT1G06440
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.276 |
0.069 |
| 444 |
251105_at |
AT5G01730
|
ATSCAR4, SCAR4, SCAR family protein 4, WAVE3 |
-0.275 |
0.049 |
| 445 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.275 |
-0.299 |
| 446 |
265632_at |
AT2G14290
|
unknown |
-0.274 |
0.766 |
| 447 |
261921_at |
AT1G65900
|
unknown |
-0.274 |
-0.205 |
| 448 |
264287_at |
AT1G61930
|
unknown |
-0.273 |
-0.188 |
| 449 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
-0.272 |
0.346 |
| 450 |
245183_at |
AT5G12440
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.271 |
-0.275 |
| 451 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.271 |
-0.052 |
| 452 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.271 |
-0.005 |
| 453 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.271 |
-0.069 |
| 454 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
-0.270 |
-0.017 |
| 455 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.269 |
0.029 |
| 456 |
266886_at |
AT2G44745
|
AtWRKY12, WRKY12, WRKY DNA-binding protein 12 |
-0.268 |
-0.053 |
| 457 |
253832_at |
AT4G27654
|
unknown |
-0.268 |
1.119 |
| 458 |
245254_at |
AT4G14680
|
APS3 |
-0.268 |
-0.010 |
| 459 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
-0.267 |
0.068 |
| 460 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
-0.266 |
0.139 |
| 461 |
246230_at |
AT4G36710
|
AtHAM4, Arabidopsis thaliana HAIRY MERISTEM 4, HAM4 |
-0.265 |
-0.114 |
| 462 |
267515_at |
AT2G45680
|
TCP9, TCP domain protein 9 |
-0.265 |
-0.039 |
| 463 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.264 |
-0.021 |
| 464 |
259400_at |
AT1G17750
|
AtPEPR2, PEP1 RECEPTOR 2, PEPR2, PEP1 receptor 2 |
-0.264 |
0.593 |
| 465 |
249972_at |
AT5G19040
|
ATIPT5, Arabidopsis thaliana ISOPENTENYLTRANSFERASE 5, IPT5, isopentenyltransferase 5 |
-0.264 |
-0.013 |
| 466 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.263 |
0.016 |
| 467 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.263 |
-0.235 |
| 468 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.263 |
-0.197 |
| 469 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.261 |
0.487 |
| 470 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
-0.261 |
0.412 |
| 471 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
-0.259 |
-0.185 |
| 472 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.259 |
-0.293 |
| 473 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.259 |
-0.012 |
| 474 |
252240_at |
AT3G50010
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.258 |
-0.013 |
| 475 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.258 |
0.413 |
| 476 |
260363_at |
AT1G70550
|
unknown |
-0.258 |
-0.371 |
| 477 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.257 |
0.293 |
| 478 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.256 |
0.715 |
| 479 |
253656_at |
AT4G30090
|
unknown |
-0.256 |
0.201 |
| 480 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
-0.255 |
0.243 |
| 481 |
266682_at |
AT2G19780
|
[Leucine-rich repeat (LRR) family protein] |
-0.255 |
0.045 |
| 482 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.255 |
-0.139 |
| 483 |
248928_at |
AT5G45970
|
ARAC2, Arabidopsis RAC-like 2, ATRAC2, ARABIDOPSIS THALIANA RAC 2, ATROP7, RHO-RELATED PROTEIN FROM PLANTS 7, RAC2, RAC-like 2, ROP7 |
-0.254 |
-0.070 |
| 484 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
-0.254 |
0.705 |
| 485 |
252651_at |
AT3G44700
|
unknown |
-0.254 |
0.033 |
| 486 |
247775_at |
AT5G58690
|
ATPLC5, PLC5, phosphatidylinositol-speciwc phospholipase C5 |
-0.254 |
-0.053 |
| 487 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
-0.254 |
-0.093 |
| 488 |
247648_at |
AT5G60020
|
ATLAC17, LAC17, laccase 17 |
-0.253 |
-0.094 |
| 489 |
264517_at |
AT1G10120
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.253 |
-0.322 |
| 490 |
265646_at |
AT2G27360
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.253 |
-0.231 |
| 491 |
251093_at |
AT5G01360
|
TBL3, TRICHOME BIREFRINGENCE-LIKE 3 |
-0.253 |
-0.095 |
| 492 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.253 |
-0.025 |
| 493 |
263688_at |
AT1G26920
|
unknown |
-0.251 |
0.164 |
| 494 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
-0.249 |
-0.464 |
| 495 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.249 |
0.395 |
| 496 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.249 |
-0.117 |
| 497 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.249 |
0.876 |
| 498 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.249 |
0.601 |
| 499 |
256968_at |
AT3G21070
|
ATNADK-1, NAD KINASE 1, NADK1, NAD kinase 1 |
-0.249 |
0.345 |
| 500 |
250246_at |
AT5G13700
|
APAO, ATPAO1, polyamine oxidase 1, PAO1, polyamine oxidase 1 |
-0.249 |
0.267 |
| 501 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.248 |
-0.113 |
| 502 |
246783_at |
AT5G27360
|
SFP2 |
-0.248 |
-0.373 |
| 503 |
265083_at |
AT1G03820
|
unknown |
-0.248 |
0.405 |
| 504 |
254562_at |
AT4G19230
|
CYP707A1, cytochrome P450, family 707, subfamily A, polypeptide 1 |
-0.248 |
0.297 |
| 505 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
-0.248 |
0.761 |
| 506 |
264363_at |
AT1G03170
|
FAF2, FANTASTIC FOUR 2, FTM5, FLORAL TRANSITION AT THE MERISTEM5 |
-0.247 |
-0.001 |
| 507 |
266481_at |
AT2G31070
|
TCP10, TCP domain protein 10 |
-0.247 |
-0.208 |
| 508 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.247 |
-0.151 |
| 509 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.247 |
0.300 |
| 510 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
-0.247 |
0.449 |
| 511 |
264324_at |
AT1G04160
|
ATXIB, ARABIDOPSIS THALIANA MYOSIN XI B, XI-8, MYOSIN XI-8, XI-B, MYOSIN XI B, XIB, myosin XI B |
-0.246 |
-0.115 |
| 512 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-0.245 |
0.422 |
| 513 |
261675_at |
AT1G18290
|
unknown |
-0.245 |
0.025 |
| 514 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.245 |
0.001 |
| 515 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.245 |
1.437 |
| 516 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.244 |
-0.168 |
| 517 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.243 |
0.042 |
| 518 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.242 |
1.018 |
| 519 |
262996_at |
AT1G54440
|
[Polynucleotidyl transferase, ribonuclease H fold protein with HRDC domain] |
-0.242 |
-0.121 |
| 520 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.241 |
-0.671 |
| 521 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.241 |
-0.503 |
| 522 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.241 |
-0.080 |
| 523 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
-0.240 |
-0.288 |
| 524 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
-0.239 |
0.304 |
| 525 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.238 |
-0.044 |
| 526 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
-0.238 |
0.213 |
| 527 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
-0.238 |
0.382 |
| 528 |
267177_at |
AT2G37580
|
[RING/U-box superfamily protein] |
-0.237 |
0.574 |
| 529 |
256021_at |
AT1G58270
|
ZW9 |
-0.237 |
0.411 |
| 530 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
-0.236 |
-0.205 |
| 531 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.236 |
0.427 |
| 532 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
-0.236 |
0.640 |
| 533 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.235 |
-0.020 |
| 534 |
249312_at |
AT5G41550
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.235 |
-0.025 |
| 535 |
260086_at |
AT1G63240
|
unknown |
-0.235 |
-0.014 |
| 536 |
264056_at |
AT2G28510
|
[Dof-type zinc finger DNA-binding family protein] |
-0.235 |
0.340 |
| 537 |
247048_at |
AT5G66560
|
[Phototropic-responsive NPH3 family protein] |
-0.234 |
-0.112 |
| 538 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.234 |
-0.418 |
| 539 |
246495_at |
AT5G16200
|
[50S ribosomal protein-related] |
-0.234 |
0.125 |
| 540 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
-0.233 |
0.592 |
| 541 |
265204_at |
AT2G36650
|
unknown |
-0.233 |
0.252 |
| 542 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
-0.233 |
0.481 |
| 543 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
-0.232 |
0.057 |
| 544 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-0.231 |
0.170 |
| 545 |
260227_at |
AT1G74450
|
unknown |
-0.231 |
0.222 |
| 546 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
-0.231 |
-0.125 |
| 547 |
248506_at |
AT5G50370
|
[Adenylate kinase family protein] |
-0.231 |
0.194 |
| 548 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.230 |
0.204 |
| 549 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.230 |
-0.110 |
| 550 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
-0.230 |
-0.018 |
| 551 |
261476_at |
AT1G14480
|
[Ankyrin repeat family protein] |
-0.229 |
0.097 |
| 552 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
-0.229 |
0.185 |
| 553 |
263795_at |
AT2G24610
|
ATCNGC14, cyclic nucleotide-gated channel 14, CNGC14, cyclic nucleotide-gated channel 14 |
-0.229 |
0.033 |
| 554 |
258540_at |
AT3G06990
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.228 |
-0.086 |
| 555 |
253123_at |
AT4G36000
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.228 |
0.111 |
| 556 |
266022_at |
AT2G05920
|
[Subtilase family protein] |
-0.228 |
-0.316 |
| 557 |
257364_at |
AT2G45940
|
unknown |
-0.227 |
0.316 |
| 558 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
-0.227 |
0.929 |
| 559 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.226 |
-0.040 |
| 560 |
252536_at |
AT3G45700
|
[Major facilitator superfamily protein] |
-0.226 |
0.021 |
| 561 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.226 |
-0.422 |
| 562 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.225 |
-0.612 |
| 563 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.225 |
-0.391 |
| 564 |
255028_at |
AT4G09890
|
unknown |
-0.225 |
-0.490 |
| 565 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
-0.224 |
0.074 |
| 566 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.224 |
1.058 |
| 567 |
257056_at |
AT3G15350
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.224 |
0.060 |
| 568 |
249903_at |
AT5G22690
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.224 |
0.238 |
| 569 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.224 |
-0.062 |
| 570 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
-0.224 |
-0.070 |
| 571 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
-0.224 |
0.075 |
| 572 |
262768_at |
AT1G12990
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.223 |
0.134 |
| 573 |
255132_at |
AT4G08170
|
[Inositol 1,3,4-trisphosphate 5/6-kinase family protein] |
-0.223 |
0.448 |
| 574 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.223 |
-0.463 |
| 575 |
264862_at |
AT1G24330
|
[ARM repeat superfamily protein] |
-0.223 |
0.221 |
| 576 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.223 |
-0.012 |
| 577 |
246512_at |
AT5G15630
|
COBL4, COBRA-LIKE4, IRX6, IRREGULAR XYLEM 6 |
-0.223 |
-0.141 |
| 578 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.222 |
0.509 |
| 579 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.222 |
0.171 |
| 580 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.222 |
-0.009 |
| 581 |
267182_at |
AT2G23360
|
unknown |
-0.222 |
-0.275 |
| 582 |
261381_at |
AT1G05460
|
SDE3, SILENCING DEFECTIVE |
-0.221 |
-0.156 |
| 583 |
246425_at |
AT5G17420
|
ATCESA7, CESA7, CELLULOSE SYNTHASE CATALYTIC SUBUNIT 7, IRX3, IRREGULAR XYLEM 3, MUR10, MURUS 10 |
-0.221 |
-0.195 |
| 584 |
256706_at |
AT3G30300
|
[O-fucosyltransferase family protein] |
-0.221 |
-0.274 |
| 585 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.220 |
0.092 |
| 586 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.220 |
0.078 |
| 587 |
256816_at |
AT3G21400
|
unknown |
-0.220 |
-0.046 |
| 588 |
250086_at |
AT5G17260
|
anac086, NAC domain containing protein 86, NAC086, NAC domain containing protein 86 |
-0.220 |
-0.040 |
| 589 |
259499_at |
AT1G15730
|
[Cobalamin biosynthesis CobW-like protein] |
-0.219 |
-0.104 |
| 590 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
-0.219 |
0.300 |
| 591 |
258860_at |
AT3G02050
|
ATKT4, ARABIDOPSIS THALIANA POTASSIUM TRANSPORTER 4, ATKUP3, KUP3, K+ uptake transporter 3 |
-0.219 |
-0.214 |
| 592 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.219 |
0.602 |
| 593 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
-0.219 |
-0.076 |
| 594 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.218 |
0.137 |
| 595 |
253830_at |
AT4G27652
|
unknown |
-0.218 |
0.727 |
| 596 |
253405_at |
AT4G32800
|
[Integrase-type DNA-binding superfamily protein] |
-0.218 |
0.360 |
| 597 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
-0.218 |
1.034 |
| 598 |
260267_at |
AT1G68530
|
CER6, ECERIFERUM 6, CUT1, CUTICULAR 1, G2, KCS6, 3-ketoacyl-CoA synthase 6, POP1, POLLEN-PISTIL INCOMPATIBILITY 1 |
-0.218 |
-0.025 |
| 599 |
254670_at |
AT4G18390
|
TCP2, TCP2, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 2 |
-0.217 |
-0.008 |
| 600 |
251995_at |
AT3G52940
|
ELL1, EXTRA-LONG-LIFESPAN 1, FK, FACKEL, HYD2 |
-0.217 |
-0.289 |