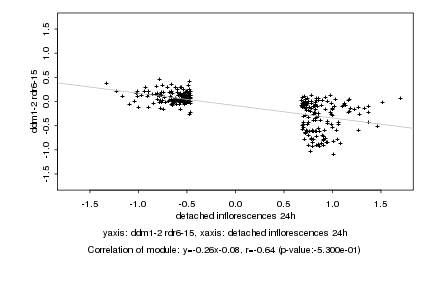
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
detached inflorescences 24h |
Log2 signal ratio
ddm1-2 rdr6-15 |
| 1 |
258327_at |
AT3G22640
|
PAP85 |
1.697 |
0.070 |
| 2 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.513 |
-0.015 |
| 3 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.462 |
-0.499 |
| 4 |
260668_at |
AT1G19530
|
unknown |
1.370 |
-0.092 |
| 5 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
1.368 |
-0.209 |
| 6 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
1.362 |
-0.423 |
| 7 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
1.321 |
-0.128 |
| 8 |
263475_at |
AT2G31945
|
unknown |
1.276 |
-0.255 |
| 9 |
249454_at |
AT5G39520
|
unknown |
1.262 |
-0.589 |
| 10 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
1.258 |
-0.114 |
| 11 |
264238_at |
AT1G54740
|
unknown |
1.219 |
-0.164 |
| 12 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
1.184 |
-0.126 |
| 13 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
1.170 |
0.052 |
| 14 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
1.169 |
-0.201 |
| 15 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
1.162 |
0.038 |
| 16 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
1.157 |
-0.217 |
| 17 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
1.126 |
-0.076 |
| 18 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
1.123 |
-0.025 |
| 19 |
254954_at |
AT4G10910
|
unknown |
1.111 |
-0.074 |
| 20 |
256109_at |
AT1G16950
|
unknown |
1.100 |
-0.090 |
| 21 |
259464_at |
AT1G18990
|
unknown |
1.080 |
-0.863 |
| 22 |
263215_at |
AT1G30710
|
[FAD-binding Berberine family protein] |
1.055 |
-0.422 |
| 23 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
1.055 |
-0.459 |
| 24 |
252881_at |
AT4G39610
|
unknown |
1.050 |
-0.769 |
| 25 |
267531_at |
AT2G41860
|
CPK14, calcium-dependent protein kinase 14 |
1.041 |
-0.580 |
| 26 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.023 |
0.043 |
| 27 |
260513_at |
AT1G51490
|
BGLU36, beta glucosidase 36 |
1.015 |
-0.101 |
| 28 |
266558_at |
AT2G23900
|
[Pectin lyase-like superfamily protein] |
1.006 |
-1.080 |
| 29 |
246123_at |
AT5G20390
|
[Glycosyl hydrolase superfamily protein] |
1.001 |
-0.807 |
| 30 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.998 |
-0.156 |
| 31 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.996 |
-0.270 |
| 32 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.990 |
-0.532 |
| 33 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
0.990 |
-0.131 |
| 34 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
0.985 |
-0.031 |
| 35 |
258600_at |
AT3G02810
|
LIP2, LOST IN POLLEN TUBE GUIDANCE 2 |
0.981 |
-0.470 |
| 36 |
258337_at |
AT3G16040
|
[Translation machinery associated TMA7] |
0.980 |
-0.260 |
| 37 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.976 |
0.125 |
| 38 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.970 |
-0.221 |
| 39 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.948 |
0.005 |
| 40 |
249194_at |
AT5G42490
|
[ATP binding microtubule motor family protein] |
0.942 |
-0.447 |
| 41 |
245036_at |
AT2G26410
|
Iqd4, IQ-domain 4 |
0.942 |
-0.847 |
| 42 |
258012_at |
AT3G19310
|
[PLC-like phosphodiesterases superfamily protein] |
0.940 |
-0.404 |
| 43 |
261045_at |
AT1G01310
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.933 |
-0.844 |
| 44 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.922 |
-0.155 |
| 45 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.920 |
0.100 |
| 46 |
263043_at |
AT1G23350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.918 |
-0.759 |
| 47 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.909 |
-0.580 |
| 48 |
257082_at |
AT3G20580
|
COBL10, COBRA-like protein 10 precursor |
0.907 |
-0.803 |
| 49 |
259820_at |
AT1G66210
|
[Subtilisin-like serine endopeptidase family protein] |
0.902 |
-0.708 |
| 50 |
257119_at |
AT3G20190
|
PRK4, pollen receptor like kinase 4 |
0.901 |
-0.899 |
| 51 |
261528_at |
AT1G14420
|
AT59 |
0.899 |
-0.781 |
| 52 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.897 |
0.035 |
| 53 |
267398_at |
AT2G44560
|
AtGH9B11, glycosyl hydrolase 9B11, GH9B11, glycosyl hydrolase 9B11 |
0.888 |
-0.684 |
| 54 |
247754_at |
AT5G59080
|
unknown |
0.885 |
-0.074 |
| 55 |
258753_at |
AT3G09530
|
ATEXO70H3, exocyst subunit exo70 family protein H3, EXO70H3, exocyst subunit exo70 family protein H3 |
0.880 |
-0.859 |
| 56 |
258168_at |
AT3G21570
|
unknown |
0.868 |
-0.382 |
| 57 |
264813_at |
AT2G17890
|
CPK16, calcium-dependent protein kinase 16 |
0.859 |
-0.602 |
| 58 |
258605_at |
AT3G02970
|
EXL6, EXORDIUM like 6 |
0.857 |
-0.920 |
| 59 |
251433_at |
AT3G59830
|
[Integrin-linked protein kinase family] |
0.855 |
-0.309 |
| 60 |
262696_at |
AT1G75870
|
unknown |
0.853 |
-0.558 |
| 61 |
267476_at |
AT2G02720
|
[Pectate lyase family protein] |
0.851 |
-0.884 |
| 62 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.851 |
0.040 |
| 63 |
263781_at |
AT2G46360
|
unknown |
0.850 |
0.067 |
| 64 |
246251_at |
AT4G37220
|
[Cold acclimation protein WCOR413 family] |
0.850 |
0.026 |
| 65 |
255733_at |
AT1G25400
|
unknown |
0.848 |
-0.228 |
| 66 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
0.845 |
-0.087 |
| 67 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.834 |
0.065 |
| 68 |
249447_at |
AT5G39400
|
ATPTEN1, PTEN1, Phosphatase and TENsin homolog deleted on chromosome ten 1 |
0.834 |
-0.713 |
| 69 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.829 |
0.025 |
| 70 |
252710_at |
AT3G43860
|
AtGH9A4, glycosyl hydrolase 9A4, GH9A4, glycosyl hydrolase 9A4 |
0.829 |
-0.899 |
| 71 |
257358_at |
AT2G40990
|
[DHHC-type zinc finger family protein] |
0.826 |
-0.317 |
| 72 |
257886_at |
AT3G17060
|
[Pectin lyase-like superfamily protein] |
0.826 |
-0.619 |
| 73 |
248086_at |
AT5G55490
|
ATGEX1, GAMETE EXPRESSED PROTEIN 1, GEX1, gamete expressed protein 1 |
0.825 |
-0.154 |
| 74 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
0.824 |
-0.063 |
| 75 |
259177_at |
AT3G01630
|
[Major facilitator superfamily protein] |
0.822 |
-0.245 |
| 76 |
255006_at |
AT4G10010
|
[Protein kinase superfamily protein] |
0.820 |
-0.335 |
| 77 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.819 |
-0.042 |
| 78 |
260161_at |
AT1G79860
|
ATROPGEF12, MEE64, MATERNAL EFFECT EMBRYO ARREST 64, ROPGEF12, ROP (rho of plants) guanine nucleotide exchange factor 12 |
0.817 |
-0.604 |
| 79 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.816 |
-0.391 |
| 80 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.815 |
-0.003 |
| 81 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.812 |
-0.221 |
| 82 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.806 |
-0.083 |
| 83 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.803 |
-0.084 |
| 84 |
254123_at |
AT4G24640
|
APPB1 |
0.803 |
-0.609 |
| 85 |
251180_at |
AT3G62640
|
unknown |
0.801 |
-0.783 |
| 86 |
264269_at |
AT1G60240
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.800 |
-0.382 |
| 87 |
259370_at |
AT1G69050
|
unknown |
0.796 |
-0.263 |
| 88 |
246545_at |
AT5G15110
|
[Pectate lyase family protein] |
0.794 |
-0.918 |
| 89 |
249448_at |
AT5G39420
|
cdc2cAt, CDC2C |
0.788 |
-0.617 |
| 90 |
260737_at |
AT1G17540
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
0.785 |
-0.593 |
| 91 |
253831_at |
AT4G27580
|
unknown |
0.785 |
-0.830 |
| 92 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.783 |
0.019 |
| 93 |
254178_at |
AT4G23880
|
unknown |
0.782 |
-0.125 |
| 94 |
250121_at |
AT5G16500
|
LIP1, LOST IN POLLEN TUBE GUIDANCE 1 |
0.781 |
-0.785 |
| 95 |
245057_at |
AT2G26490
|
[Transducin/WD40 repeat-like superfamily protein] |
0.778 |
-0.757 |
| 96 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
0.776 |
-0.038 |
| 97 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.774 |
0.137 |
| 98 |
263450_at |
AT2G31500
|
CPK24, calcium-dependent protein kinase 24 |
0.771 |
-1.015 |
| 99 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.764 |
-0.197 |
| 100 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.757 |
-0.108 |
| 101 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.756 |
-0.619 |
| 102 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.752 |
-0.323 |
| 103 |
257433_at |
AT2G21990
|
unknown |
0.749 |
-0.433 |
| 104 |
257090_at |
AT3G20530
|
[Protein kinase superfamily protein] |
0.748 |
-0.692 |
| 105 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
0.748 |
0.015 |
| 106 |
264635_at |
AT1G65500
|
unknown |
0.744 |
-0.174 |
| 107 |
248470_at |
AT5G50830
|
unknown |
0.743 |
-0.909 |
| 108 |
246918_at |
AT5G25340
|
[Ubiquitin-like superfamily protein] |
0.742 |
-0.466 |
| 109 |
245053_at |
AT2G26450
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.741 |
-0.607 |
| 110 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.737 |
0.068 |
| 111 |
254787_at |
AT4G12690
|
unknown |
0.733 |
-0.030 |
| 112 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.729 |
-0.043 |
| 113 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
0.728 |
-0.080 |
| 114 |
257405_at |
AT1G24620
|
[EF hand calcium-binding protein family] |
0.728 |
-0.581 |
| 115 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
0.727 |
-0.118 |
| 116 |
256105_at |
AT1G16910
|
LSH8, LIGHT SENSITIVE HYPOCOTYLS 8 |
0.727 |
-0.024 |
| 117 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.723 |
-0.182 |
| 118 |
263144_at |
AT1G54070
|
[Dormancy/auxin associated family protein] |
0.720 |
-0.654 |
| 119 |
259856_at |
AT1G68440
|
unknown |
0.719 |
0.008 |
| 120 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.716 |
-0.060 |
| 121 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
0.716 |
-0.286 |
| 122 |
247921_at |
AT5G57660
|
ATCOL5, BBX6, B-box domain protein 6, COL5, CONSTANS-like 5 |
0.715 |
-0.029 |
| 123 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.715 |
-0.141 |
| 124 |
255276_at |
AT4G04930
|
DES-1-LIKE |
0.714 |
-0.622 |
| 125 |
260011_at |
AT1G68110
|
[ENTH/ANTH/VHS superfamily protein] |
0.712 |
-0.634 |
| 126 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.712 |
0.061 |
| 127 |
252462_at |
AT3G47250
|
unknown |
0.711 |
-0.070 |
| 128 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.709 |
0.111 |
| 129 |
253356_at |
AT4G33390
|
unknown |
0.704 |
-0.106 |
| 130 |
258889_at |
AT3G05610
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.704 |
-0.779 |
| 131 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.698 |
-0.014 |
| 132 |
265923_at |
AT2G18470
|
AtPERK4, proline-rich extensin-like receptor kinase 4, PERK4, proline-rich extensin-like receptor kinase 4 |
0.698 |
-0.519 |
| 133 |
255251_at |
AT4G04980
|
unknown |
0.694 |
-0.306 |
| 134 |
253034_at |
AT4G38230
|
ATCPK26, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 26, CPK26, calcium-dependent protein kinase 26 |
0.694 |
-0.432 |
| 135 |
257672_at |
AT3G20300
|
unknown |
0.693 |
-0.472 |
| 136 |
263118_at |
AT1G03090
|
MCCA |
0.693 |
-0.033 |
| 137 |
259015_at |
AT3G07350
|
unknown |
0.689 |
-0.031 |
| 138 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.688 |
0.099 |
| 139 |
246782_at |
AT5G27320
|
ATGID1C, GID1C, GA INSENSITIVE DWARF1C |
0.687 |
-0.092 |
| 140 |
247199_at |
AT5G65210
|
TGA1, TGACG sequence-specific binding protein 1 |
0.686 |
-0.112 |
| 141 |
245700_at |
AT5G04180
|
ACA3, alpha carbonic anhydrase 3, ATACA3, ALPHA CARBONIC ANHYDRASE 3 |
0.685 |
-0.575 |
| 142 |
262146_at |
AT1G52580
|
ATRBL5, RHOMBOID-like protein 5, RBL5, RHOMBOID-like protein 5 |
0.683 |
-0.494 |
| 143 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
0.682 |
-0.099 |
| 144 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.680 |
0.041 |
| 145 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.677 |
-0.064 |
| 146 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.337 |
0.389 |
| 147 |
267440_at |
AT2G19070
|
SHT, spermidine hydroxycinnamoyl transferase |
-1.227 |
0.212 |
| 148 |
259744_at |
AT1G71160
|
KCS7, 3-ketoacyl-CoA synthase 7 |
-1.168 |
0.106 |
| 149 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-1.094 |
-0.055 |
| 150 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-1.043 |
0.005 |
| 151 |
261921_at |
AT1G65900
|
unknown |
-1.029 |
0.183 |
| 152 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-1.019 |
0.217 |
| 153 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
-1.017 |
0.109 |
| 154 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-1.005 |
-0.104 |
| 155 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
-0.969 |
0.133 |
| 156 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.939 |
0.212 |
| 157 |
256125_at |
AT1G18250
|
ATLP-1 |
-0.929 |
0.296 |
| 158 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.916 |
0.117 |
| 159 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.904 |
0.226 |
| 160 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
-0.899 |
0.152 |
| 161 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.899 |
-0.121 |
| 162 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.865 |
0.146 |
| 163 |
263840_at |
AT2G36885
|
unknown |
-0.851 |
-0.035 |
| 164 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.826 |
0.157 |
| 165 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
-0.818 |
0.326 |
| 166 |
258480_at |
AT3G02640
|
unknown |
-0.813 |
0.182 |
| 167 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.812 |
0.042 |
| 168 |
255460_at |
AT4G02800
|
unknown |
-0.806 |
0.127 |
| 169 |
264363_at |
AT1G03170
|
FAF2, FANTASTIC FOUR 2, FTM5, FLORAL TRANSITION AT THE MERISTEM5 |
-0.792 |
-0.019 |
| 170 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.786 |
0.475 |
| 171 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
-0.784 |
0.107 |
| 172 |
254786_at |
AT4G12890
|
[Gamma interferon responsive lysosomal thiol (GILT) reductase family protein] |
-0.782 |
0.149 |
| 173 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.781 |
0.035 |
| 174 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.775 |
-0.142 |
| 175 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.770 |
0.187 |
| 176 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.770 |
0.141 |
| 177 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.766 |
-0.008 |
| 178 |
248139_at |
AT5G54970
|
unknown |
-0.762 |
0.037 |
| 179 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.758 |
0.343 |
| 180 |
249497_at |
AT5G39220
|
[alpha/beta-Hydrolases superfamily protein] |
-0.749 |
0.199 |
| 181 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.742 |
-0.164 |
| 182 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
-0.741 |
0.085 |
| 183 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.729 |
-0.019 |
| 184 |
246526_at |
AT5G15720
|
GLIP7, GDSL-motif lipase 7 |
-0.704 |
0.293 |
| 185 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.696 |
0.012 |
| 186 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.688 |
0.038 |
| 187 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.687 |
0.196 |
| 188 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.674 |
0.112 |
| 189 |
251920_at |
AT3G53900
|
PYRR, PYRIMIDINE R, UPP, uracil phosphoribosyltransferase |
-0.669 |
0.043 |
| 190 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.668 |
-0.055 |
| 191 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
-0.664 |
0.222 |
| 192 |
251753_at |
AT3G55760
|
unknown |
-0.663 |
0.094 |
| 193 |
251560_at |
AT3G57920
|
SPL15, squamosa promoter binding protein-like 15 |
-0.661 |
0.359 |
| 194 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
-0.657 |
0.176 |
| 195 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.653 |
-0.069 |
| 196 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
-0.651 |
0.018 |
| 197 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.649 |
-0.028 |
| 198 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.646 |
-0.032 |
| 199 |
248687_at |
AT5G48300
|
ADG1, ADP glucose pyrophosphorylase 1, APS1, ADP-GLUCOSE PYROPHOSPHORYLASE SMALL SUBUNIT 1 |
-0.644 |
0.031 |
| 200 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.640 |
-0.009 |
| 201 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.637 |
0.021 |
| 202 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
-0.631 |
0.011 |
| 203 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.631 |
-0.019 |
| 204 |
261223_at |
AT1G19950
|
HVA22H, HVA22-like protein H (ATHVA22H) |
-0.630 |
0.017 |
| 205 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
-0.621 |
0.071 |
| 206 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
-0.620 |
0.303 |
| 207 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.619 |
0.060 |
| 208 |
245884_at |
AT5G09300
|
[Thiamin diphosphate-binding fold (THDP-binding) superfamily protein] |
-0.617 |
0.054 |
| 209 |
258702_at |
AT3G09730
|
unknown |
-0.613 |
-0.059 |
| 210 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.606 |
0.202 |
| 211 |
260565_at |
AT2G43800
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.599 |
0.256 |
| 212 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.597 |
0.141 |
| 213 |
263787_at |
AT2G46420
|
unknown |
-0.597 |
0.143 |
| 214 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.594 |
0.006 |
| 215 |
248853_at |
AT5G46570
|
BSK2, brassinosteroid-signaling kinase 2 |
-0.587 |
0.023 |
| 216 |
262806_at |
AT1G20950
|
[Phosphofructokinase family protein] |
-0.586 |
0.019 |
| 217 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
-0.585 |
0.160 |
| 218 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.583 |
0.057 |
| 219 |
263287_at |
AT2G36145
|
unknown |
-0.581 |
-0.047 |
| 220 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
-0.580 |
0.033 |
| 221 |
261137_at |
AT1G19830
|
SAUR54, SMALL AUXIN UPREGULATED RNA 54 |
-0.578 |
-0.003 |
| 222 |
247425_at |
AT5G62550
|
unknown |
-0.577 |
0.049 |
| 223 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.574 |
-0.152 |
| 224 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.574 |
0.128 |
| 225 |
261642_at |
AT1G27680
|
APL2, ADPGLC-PPase large subunit |
-0.574 |
0.309 |
| 226 |
267639_at |
AT2G42200
|
AtSPL9, SPL9, squamosa promoter binding protein-like 9 |
-0.573 |
0.174 |
| 227 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.570 |
-0.010 |
| 228 |
247235_at |
AT5G64580
|
EMB3144, EMBRYO DEFECTIVE 3144 |
-0.568 |
0.020 |
| 229 |
253817_at |
AT4G28310
|
unknown |
-0.566 |
0.110 |
| 230 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.566 |
0.308 |
| 231 |
260028_at |
AT1G29980
|
unknown |
-0.559 |
0.067 |
| 232 |
250326_at |
AT5G12080
|
ATMSL10, MSL10, mechanosensitive channel of small conductance-like 10 |
-0.558 |
0.145 |
| 233 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.554 |
0.053 |
| 234 |
261898_at |
AT1G80720
|
[Mitochondrial glycoprotein family protein] |
-0.553 |
0.059 |
| 235 |
258997_at |
AT3G01810
|
unknown |
-0.550 |
0.136 |
| 236 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.550 |
0.080 |
| 237 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.549 |
0.114 |
| 238 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.548 |
0.037 |
| 239 |
253480_at |
AT4G31840
|
AtENODL15, ENODL15, early nodulin-like protein 15 |
-0.545 |
0.253 |
| 240 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.542 |
0.185 |
| 241 |
256962_at |
AT3G13560
|
[O-Glycosyl hydrolases family 17 protein] |
-0.541 |
0.117 |
| 242 |
248309_at |
AT5G52540
|
unknown |
-0.541 |
-0.001 |
| 243 |
260645_at |
AT1G53300
|
TTL1, tetratricopeptide-repeat thioredoxin-like 1 |
-0.540 |
0.042 |
| 244 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.540 |
0.045 |
| 245 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.539 |
0.046 |
| 246 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
-0.538 |
0.024 |
| 247 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.535 |
-0.056 |
| 248 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.534 |
0.130 |
| 249 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
-0.532 |
0.076 |
| 250 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.528 |
0.005 |
| 251 |
263782_at |
AT2G46380
|
unknown |
-0.527 |
-0.028 |
| 252 |
264581_at |
AT1G05210
|
[Transmembrane protein 97, predicted] |
-0.524 |
0.062 |
| 253 |
250920_at |
AT5G03390
|
unknown |
-0.524 |
0.092 |
| 254 |
247400_at |
AT5G62840
|
[Phosphoglycerate mutase family protein] |
-0.521 |
0.053 |
| 255 |
262783_at |
AT1G10850
|
[Leucine-rich repeat protein kinase family protein] |
-0.520 |
0.091 |
| 256 |
247192_at |
AT5G65360
|
H3.1, histone 3.1 |
-0.519 |
0.161 |
| 257 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.518 |
0.159 |
| 258 |
251394_at |
AT3G60900
|
FLA10, FASCICLIN-like arabinogalactan-protein 10 |
-0.518 |
0.194 |
| 259 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.516 |
-0.012 |
| 260 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.512 |
-0.008 |
| 261 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
-0.511 |
0.128 |
| 262 |
257814_at |
AT3G25110
|
AtFaTA, fatA acyl-ACP thioesterase, FaTA, fatA acyl-ACP thioesterase |
-0.511 |
-0.024 |
| 263 |
258474_at |
AT3G02650
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.511 |
0.208 |
| 264 |
253363_at |
AT4G33130
|
unknown |
-0.509 |
0.112 |
| 265 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
-0.508 |
0.066 |
| 266 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.500 |
0.062 |
| 267 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.497 |
0.009 |
| 268 |
251287_at |
AT3G61820
|
[Eukaryotic aspartyl protease family protein] |
-0.496 |
0.013 |
| 269 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.495 |
0.224 |
| 270 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
-0.493 |
0.069 |
| 271 |
264668_at |
AT1G09780
|
iPGAM1, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 1 |
-0.491 |
-0.006 |
| 272 |
247039_at |
AT5G67270
|
ATEB1C, MICROTUBULE END BINDING PROTEIN 1, ATEB1H1, ATEB1-HOMOLOG1, EB1C, end binding protein 1C |
-0.491 |
0.348 |
| 273 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
-0.490 |
0.097 |
| 274 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.488 |
0.035 |
| 275 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
-0.487 |
0.169 |
| 276 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
-0.483 |
0.178 |
| 277 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.482 |
0.070 |
| 278 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.482 |
0.142 |
| 279 |
260667_at |
AT1G19440
|
KCS4, 3-ketoacyl-CoA synthase 4 |
-0.478 |
-0.250 |
| 280 |
262121_at |
AT1G02800
|
ATCEL2, cellulase 2, CEL2, CEL2, cellulase 2 |
-0.477 |
0.261 |
| 281 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
-0.475 |
0.424 |
| 282 |
253600_at |
AT4G30810
|
scpl29, serine carboxypeptidase-like 29 |
-0.473 |
0.130 |
| 283 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.473 |
0.127 |
| 284 |
248105_at |
AT5G55280
|
ATFTSZ1-1, ARABIDOPSIS THALIANA HOMOLOG OF BACTERIAL CYTOKINESIS Z-RING PROTEIN FTSZ 1-1, CPFTSZ, CHLOROPLAST FTSZ, FTSZ1-1, homolog of bacterial cytokinesis Z-ring protein FTSZ 1-1 |
-0.473 |
-0.022 |
| 285 |
245165_at |
AT2G33180
|
unknown |
-0.472 |
-0.225 |
| 286 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
-0.472 |
0.103 |
| 287 |
256077_at |
AT1G18090
|
[5'-3' exonuclease family protein] |
-0.470 |
0.077 |
| 288 |
247427_at |
AT5G62580
|
[ARM repeat superfamily protein] |
-0.465 |
0.013 |
| 289 |
251083_at |
AT5G01590
|
TIC56, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 56 |
-0.465 |
0.221 |