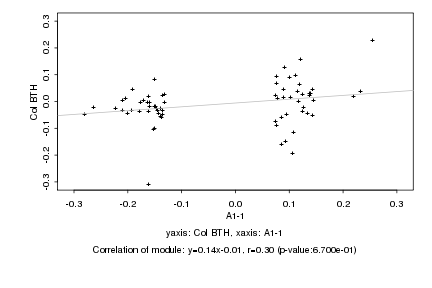
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
A1-1 |
Log2 signal ratio
Col BTH |
| 1 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
0.253 |
0.231 |
| 2 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
0.232 |
0.038 |
| 3 |
250150_at |
AT5G14710
|
unknown |
0.218 |
0.019 |
| 4 |
265083_at |
AT1G03820
|
unknown |
0.144 |
0.007 |
| 5 |
261337_at |
AT1G44810
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
0.143 |
0.045 |
| 6 |
262782_at |
AT1G13195
|
[RING/U-box superfamily protein] |
0.142 |
-0.052 |
| 7 |
250107_at |
AT5G15330
|
ATSPX4, ARABIDOPSIS THALIANA SPX DOMAIN GENE 4, SPX4, SPX domain gene 4 |
0.138 |
0.030 |
| 8 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.137 |
0.026 |
| 9 |
259463_at |
AT1G18950
|
[DDT domain superfamily] |
0.136 |
0.032 |
| 10 |
264518_at |
AT1G09980
|
[Putative serine esterase family protein] |
0.133 |
-0.042 |
| 11 |
257741_at |
AT3G27340
|
unknown |
0.126 |
-0.021 |
| 12 |
263039_at |
AT1G23280
|
[MAK16 protein-related] |
0.124 |
0.027 |
| 13 |
255952_at |
AT1G22130
|
AGL104, AGAMOUS-like 104 |
0.124 |
-0.035 |
| 14 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.120 |
0.158 |
| 15 |
249943_at |
AT5G22280
|
unknown |
0.119 |
0.064 |
| 16 |
249870_at |
AT5G23080
|
TGH, TOUGH |
0.116 |
0.002 |
| 17 |
247942_at |
AT5G57120
|
unknown |
0.114 |
0.039 |
| 18 |
256583_at |
AT3G28850
|
[Glutaredoxin family protein] |
0.111 |
0.098 |
| 19 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
0.107 |
-0.112 |
| 20 |
249256_at |
AT5G41620
|
unknown |
0.105 |
-0.193 |
| 21 |
255461_at |
AT4G02780
|
ABC33, ATCPS1, ARABIDOPSIS THALIANA ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, CPS, CPP synthase, CPS1, ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, GA1, GA REQUIRING 1 |
0.102 |
0.018 |
| 22 |
257273_at |
AT3G28030
|
UVH3, ULTRAVIOLET HYPERSENSITIVE 3, UVR1, UV REPAIR DEFECTIVE 1 |
0.099 |
0.092 |
| 23 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.094 |
-0.048 |
| 24 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.092 |
-0.149 |
| 25 |
250463_at |
AT5G10030
|
OBF4, OCS ELEMENT BINDING FACTOR 4, TGA4, TGACG motif-binding factor 4 |
0.091 |
0.130 |
| 26 |
260945_at |
AT1G05950
|
unknown |
0.088 |
0.047 |
| 27 |
265528_at |
AT2G06220
|
unknown |
0.088 |
0.017 |
| 28 |
265498_at |
AT2G15740
|
[C2H2-like zinc finger protein] |
0.085 |
-0.056 |
| 29 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
0.084 |
-0.160 |
| 30 |
247440_at |
AT5G62680
|
AtNPF2.11, GTR2, GLUCOSINOLATE TRANSPORTER-2, NPF2.11, NRT1/ PTR family 2.11 |
0.077 |
0.014 |
| 31 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
0.076 |
-0.089 |
| 32 |
265460_at |
AT2G46600
|
[Calcium-binding EF-hand family protein] |
0.076 |
0.094 |
| 33 |
267467_at |
AT2G30580
|
BMI1A, DRIP2, DREB2A-interacting protein 2 |
0.075 |
0.070 |
| 34 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.073 |
-0.072 |
| 35 |
265975_at |
AT2G11270
|
[citrate synthase-related] |
0.073 |
0.025 |
| 36 |
261040_at |
AT1G17370
|
UBP1B, oligouridylate binding protein 1B |
-0.281 |
-0.048 |
| 37 |
250223_at |
AT5G14070
|
ROXY2 |
-0.265 |
-0.019 |
| 38 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
-0.223 |
-0.023 |
| 39 |
261961_at |
AT1G36510
|
[Nucleic acid-binding proteins superfamily] |
-0.211 |
-0.030 |
| 40 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
-0.210 |
0.005 |
| 41 |
247053_at |
AT5G66710
|
[Protein kinase superfamily protein] |
-0.205 |
0.014 |
| 42 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.201 |
-0.042 |
| 43 |
260634_at |
AT1G62410
|
[MIF4G domain-containing protein] |
-0.194 |
-0.032 |
| 44 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
-0.193 |
0.046 |
| 45 |
253134_at |
AT4G35820
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.179 |
-0.037 |
| 46 |
248991_at |
AT5G45220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.177 |
-0.000 |
| 47 |
260689_at |
AT1G32290
|
unknown |
-0.171 |
0.006 |
| 48 |
257278_at |
AT3G14480
|
[glycine/proline-rich protein] |
-0.164 |
-0.001 |
| 49 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.163 |
-0.306 |
| 50 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.163 |
0.021 |
| 51 |
257179_at |
AT3G13170
|
ATSPO11-1 |
-0.163 |
-0.035 |
| 52 |
245615_at |
AT4G14470
|
[non-LTR retrotransposon family (LINE), has a 2.1e-25 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-0.161 |
-0.018 |
| 53 |
257752_at |
AT3G18720
|
[F-box family protein] |
-0.161 |
-0.002 |
| 54 |
258704_at |
AT3G09780
|
ATCRR1, CCR1, CRINKLY4 related 1 |
-0.153 |
-0.103 |
| 55 |
261987_at |
AT1G33710
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
-0.152 |
-0.017 |
| 56 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
-0.151 |
0.083 |
| 57 |
262106_at |
AT1G02970
|
ATWEE1, WEE1, WEE1 kinase homolog |
-0.151 |
-0.097 |
| 58 |
256135_at |
AT1G48730
|
unknown |
-0.149 |
-0.015 |
| 59 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
-0.147 |
-0.024 |
| 60 |
252251_at |
AT3G49820
|
unknown |
-0.146 |
-0.033 |
| 61 |
257449_at |
AT2G31420
|
[FUNCTIONS IN: DNA binding] |
-0.143 |
-0.042 |
| 62 |
259453_at |
AT1G44090
|
ATGA20OX5, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 5, GA20OX5, gibberellin 20-oxidase 5 |
-0.140 |
-0.024 |
| 63 |
258664_at |
AT3G08700
|
UBC12, ubiquitin-conjugating enzyme 12 |
-0.140 |
-0.053 |
| 64 |
247953_at |
AT5G57260
|
CYP71B10, cytochrome P450, family 71, subfamily B, polypeptide 10 |
-0.138 |
-0.057 |
| 65 |
251878_at |
AT3G54310
|
unknown |
-0.137 |
-0.047 |
| 66 |
254070_at |
AT4G25430
|
TRM23, TON1 Recruiting Motif 23 |
-0.136 |
0.023 |
| 67 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.136 |
-0.030 |
| 68 |
247050_at |
AT5G66660
|
unknown |
-0.133 |
-0.003 |
| 69 |
247868_at |
AT5G57620
|
AtMYB36, myb domain protein 36, MYB36, myb domain protein 36 |
-0.133 |
0.029 |