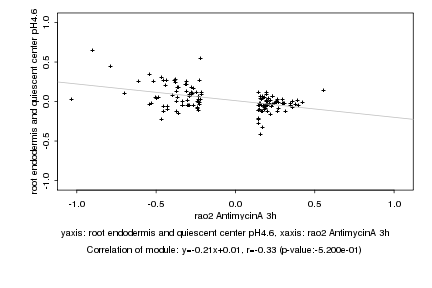
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rao2 AntimycinA 3h |
Log2 signal ratio
root endodermis and quiescent center pH4.6 |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.553 |
0.142 |
| 2 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
0.422 |
-0.003 |
| 3 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.395 |
-0.038 |
| 4 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.388 |
0.022 |
| 5 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.376 |
-0.028 |
| 6 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.358 |
-0.071 |
| 7 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.355 |
0.011 |
| 8 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.347 |
-0.020 |
| 9 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.343 |
-0.039 |
| 10 |
262384_at |
AT1G72950
|
[Disease resistance protein (TIR-NBS class)] |
0.311 |
-0.118 |
| 11 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.307 |
-0.020 |
| 12 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.298 |
-0.019 |
| 13 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.296 |
0.030 |
| 14 |
256141_at |
AT1G48640
|
[Transmembrane amino acid transporter family protein] |
0.291 |
-0.025 |
| 15 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
0.270 |
-0.012 |
| 16 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.269 |
-0.082 |
| 17 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.264 |
-0.123 |
| 18 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.261 |
0.031 |
| 19 |
257057_at |
AT3G15310
|
unknown |
0.257 |
0.003 |
| 20 |
252873_at |
AT4G40020
|
[Myosin heavy chain-related protein] |
0.253 |
-0.012 |
| 21 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.248 |
-0.004 |
| 22 |
265619_at |
AT2G27320
|
unknown |
0.246 |
-0.020 |
| 23 |
253655_at |
AT4G30070
|
LCR59, low-molecular-weight cysteine-rich 59 |
0.233 |
0.067 |
| 24 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.230 |
0.075 |
| 25 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.229 |
-0.028 |
| 26 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.223 |
-0.063 |
| 27 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.221 |
-0.062 |
| 28 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
0.219 |
0.020 |
| 29 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.217 |
-0.158 |
| 30 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
0.209 |
-0.020 |
| 31 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
0.206 |
0.049 |
| 32 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
0.201 |
-0.014 |
| 33 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.201 |
-0.047 |
| 34 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.200 |
-0.121 |
| 35 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.197 |
0.002 |
| 36 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
0.195 |
0.126 |
| 37 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.195 |
0.011 |
| 38 |
252346_at |
AT3G48650
|
[pseudogene] |
0.195 |
0.096 |
| 39 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
0.190 |
-0.031 |
| 40 |
254647_at |
AT4G18540
|
unknown |
0.190 |
0.013 |
| 41 |
264646_at |
AT1G08860
|
BON3, BONZAI 3 |
0.189 |
-0.086 |
| 42 |
259952_at |
AT1G71400
|
AtRLP12, receptor like protein 12, RLP12, receptor like protein 12 |
0.188 |
-0.051 |
| 43 |
253002_at |
AT4G38530
|
ATPLC1, phospholipase C1, PLC1, phospholipase C1 |
0.181 |
-0.075 |
| 44 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.180 |
-0.089 |
| 45 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.180 |
0.063 |
| 46 |
265539_at |
AT2G15830
|
unknown |
0.178 |
-0.056 |
| 47 |
265892_at |
AT2G15020
|
unknown |
0.177 |
-0.059 |
| 48 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.171 |
-0.046 |
| 49 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.170 |
0.043 |
| 50 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.170 |
-0.121 |
| 51 |
264528_at |
AT1G30810
|
JMJ18, Jumonji domain-containing protein 18 |
0.168 |
0.067 |
| 52 |
258091_at |
AT3G14560
|
unknown |
0.166 |
-0.321 |
| 53 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
0.164 |
-0.041 |
| 54 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
0.164 |
0.030 |
| 55 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.162 |
-0.124 |
| 56 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.156 |
0.048 |
| 57 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.154 |
0.072 |
| 58 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.153 |
-0.014 |
| 59 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
0.152 |
-0.413 |
| 60 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.151 |
-0.093 |
| 61 |
255016_at |
AT4G10120
|
ATSPS4F |
0.148 |
-0.045 |
| 62 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
0.146 |
-0.023 |
| 63 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
0.145 |
0.121 |
| 64 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.145 |
-0.223 |
| 65 |
255028_at |
AT4G09890
|
unknown |
0.145 |
-0.214 |
| 66 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.144 |
-0.104 |
| 67 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
0.143 |
-0.277 |
| 68 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.143 |
-0.101 |
| 69 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.142 |
-0.040 |
| 70 |
251871_at |
AT3G54520
|
unknown |
-1.038 |
0.028 |
| 71 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.906 |
0.654 |
| 72 |
259947_at |
AT1G71530
|
[Protein kinase superfamily protein] |
-0.792 |
0.450 |
| 73 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.702 |
0.103 |
| 74 |
266618_at |
AT2G35480
|
unknown |
-0.617 |
0.258 |
| 75 |
266606_at |
AT2G46310
|
CRF5, cytokinin response factor 5 |
-0.546 |
-0.030 |
| 76 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.544 |
0.349 |
| 77 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.534 |
-0.024 |
| 78 |
265422_at |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
-0.522 |
0.259 |
| 79 |
260848_at |
AT1G21850
|
sks8, SKU5 similar 8 |
-0.507 |
0.052 |
| 80 |
258216_at |
AT3G17980
|
AtC2, Arabidopsis thaliana C2 domain, AtGAP1, homolog of OsGAP1, C2, C2 domain |
-0.498 |
0.040 |
| 81 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
-0.489 |
0.055 |
| 82 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
-0.472 |
0.312 |
| 83 |
245710_at |
AT5G04330
|
CYP84A4, CYTOCHROME P450 84A4 |
-0.467 |
-0.219 |
| 84 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
-0.458 |
0.269 |
| 85 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
-0.458 |
-0.116 |
| 86 |
254055_at |
AT4G25330
|
unknown |
-0.455 |
-0.057 |
| 87 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
-0.443 |
0.207 |
| 88 |
263151_at |
AT1G54120
|
unknown |
-0.435 |
0.271 |
| 89 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
-0.433 |
-0.056 |
| 90 |
266738_at |
AT2G47010
|
unknown |
-0.429 |
-0.094 |
| 91 |
247287_at |
AT5G64230
|
unknown |
-0.403 |
0.081 |
| 92 |
252539_at |
AT3G45730
|
unknown |
-0.389 |
0.269 |
| 93 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.381 |
0.247 |
| 94 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
-0.378 |
0.285 |
| 95 |
245431_at |
AT4G17080
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
-0.377 |
0.002 |
| 96 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.377 |
-0.119 |
| 97 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
-0.374 |
0.138 |
| 98 |
257184_at |
AT3G13090
|
ABCC6, ATP-binding cassette C6, ATMRP8, multidrug resistance-associated protein 8, MRP8, multidrug resistance-associated protein 8 |
-0.369 |
0.060 |
| 99 |
253824_at |
AT4G27940
|
ATMTM1, ARABIDOPSIS MANGANESE TRACKING FACTOR FOR MITOCHONDRIAL SOD2, MTM1, manganese tracking factor for mitochondrial SOD2 |
-0.369 |
0.186 |
| 100 |
247176_at |
AT5G65110
|
ACX2, acyl-CoA oxidase 2, ATACX2 |
-0.364 |
0.182 |
| 101 |
263734_at |
AT1G60030
|
ATNAT7, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 7, NAT7, nucleobase-ascorbate transporter 7 |
-0.360 |
-0.141 |
| 102 |
250821_at |
AT5G05190
|
unknown |
-0.338 |
-0.049 |
| 103 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
-0.337 |
0.004 |
| 104 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
-0.337 |
-0.038 |
| 105 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.334 |
-0.038 |
| 106 |
253796_at |
AT4G28460
|
unknown |
-0.321 |
0.222 |
| 107 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
-0.314 |
0.261 |
| 108 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.313 |
0.221 |
| 109 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
-0.311 |
0.134 |
| 110 |
247913_at |
AT5G57510
|
unknown |
-0.305 |
0.014 |
| 111 |
265003_at |
AT1G26970
|
[Protein kinase superfamily protein] |
-0.303 |
-0.046 |
| 112 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.296 |
-0.049 |
| 113 |
247989_at |
AT5G56350
|
[Pyruvate kinase family protein] |
-0.294 |
0.098 |
| 114 |
259340_at |
AT3G03870
|
unknown |
-0.293 |
-0.049 |
| 115 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
-0.292 |
0.075 |
| 116 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
-0.282 |
0.101 |
| 117 |
258679_at |
AT3G08590
|
iPGAM2, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 2 |
-0.280 |
0.125 |
| 118 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
-0.279 |
0.182 |
| 119 |
265192_at |
AT1G05060
|
unknown |
-0.274 |
0.107 |
| 120 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
-0.268 |
0.173 |
| 121 |
245999_at |
AT5G20650
|
COPT5, copper transporter 5 |
-0.268 |
-0.047 |
| 122 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
-0.246 |
0.120 |
| 123 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
-0.242 |
-0.081 |
| 124 |
255647_at |
AT4G00900
|
ATECA2, ARABIDOPSIS THALIANA ER-TYPE CA2+-ATPASE 2, ECA2, ER-type Ca2+-ATPase 2 |
-0.240 |
-0.068 |
| 125 |
245439_at |
AT4G16670
|
unknown |
-0.240 |
0.006 |
| 126 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
-0.239 |
0.012 |
| 127 |
245796_at |
AT1G32230
|
ATP8, ARABIDOPSIS THALIANA P8 (INTERACTING PROTEIN), AtRCD1, CEO, CEO1, RCD1, RADICAL-INDUCED CELL DEATH1 |
-0.239 |
0.030 |
| 128 |
254603_at |
AT4G19045
|
[Mob1/phocein family protein] |
-0.238 |
-0.109 |
| 129 |
259969_at |
AT1G76550
|
[Phosphofructokinase family protein] |
-0.235 |
-0.008 |
| 130 |
259222_at |
AT3G03680
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.235 |
0.068 |
| 131 |
262219_at |
AT1G74750
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.232 |
0.275 |
| 132 |
261063_at |
AT1G07520
|
[GRAS family transcription factor] |
-0.229 |
-0.037 |
| 133 |
262514_at |
AT1G34190
|
anac017, NAC domain containing protein 17, NAC017, NAC domain containing protein 17 |
-0.228 |
-0.001 |
| 134 |
254352_at |
AT4G22320
|
unknown |
-0.225 |
0.028 |
| 135 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.221 |
0.549 |
| 136 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.219 |
0.101 |
| 137 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.215 |
0.115 |