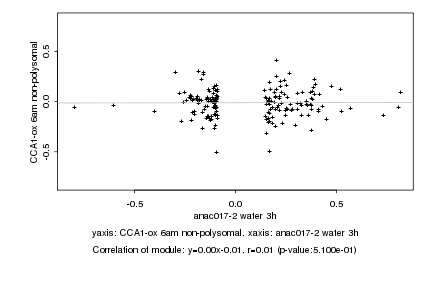
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
anac017-2 water 3h |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.816 |
0.091 |
| 2 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.807 |
-0.057 |
| 3 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.733 |
-0.139 |
| 4 |
263515_at |
AT2G21640
|
unknown |
0.567 |
-0.067 |
| 5 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.524 |
-0.098 |
| 6 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.519 |
0.123 |
| 7 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.473 |
0.150 |
| 8 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.448 |
-0.178 |
| 9 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.429 |
-0.044 |
| 10 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.415 |
0.076 |
| 11 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.410 |
-0.098 |
| 12 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.408 |
-0.076 |
| 13 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.394 |
0.179 |
| 14 |
260522_x_at |
AT2G41730
|
unknown |
0.390 |
0.225 |
| 15 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.388 |
0.071 |
| 16 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.386 |
0.147 |
| 17 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.381 |
0.025 |
| 18 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.379 |
0.105 |
| 19 |
246584_at |
AT5G14730
|
unknown |
0.373 |
-0.078 |
| 20 |
266618_at |
AT2G35480
|
unknown |
0.373 |
-0.287 |
| 21 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
0.373 |
0.034 |
| 22 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.372 |
-0.035 |
| 23 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.370 |
-0.023 |
| 24 |
250182_at |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
0.368 |
0.098 |
| 25 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.362 |
-0.139 |
| 26 |
263182_at |
AT1G05575
|
unknown |
0.353 |
-0.034 |
| 27 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.352 |
0.007 |
| 28 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.351 |
-0.029 |
| 29 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.328 |
0.099 |
| 30 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.325 |
-0.135 |
| 31 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.323 |
-0.030 |
| 32 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.321 |
-0.018 |
| 33 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.307 |
-0.015 |
| 34 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.306 |
-0.072 |
| 35 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.304 |
0.086 |
| 36 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.297 |
-0.229 |
| 37 |
249377_at |
AT5G40690
|
unknown |
0.281 |
-0.078 |
| 38 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.273 |
-0.081 |
| 39 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.272 |
-0.024 |
| 40 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.265 |
0.285 |
| 41 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.257 |
0.043 |
| 42 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.256 |
-0.075 |
| 43 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.252 |
0.161 |
| 44 |
263165_at |
AT1G03060
|
SPI, SPIRRIG |
0.249 |
-0.062 |
| 45 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.248 |
-0.083 |
| 46 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
0.244 |
-0.130 |
| 47 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.242 |
0.078 |
| 48 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.242 |
0.217 |
| 49 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.232 |
-0.218 |
| 50 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.228 |
0.099 |
| 51 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.225 |
-0.011 |
| 52 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.223 |
0.158 |
| 53 |
254289_at |
AT4G22980
|
unknown |
0.221 |
0.208 |
| 54 |
247223_at |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
0.215 |
0.040 |
| 55 |
261748_at |
AT1G76070
|
unknown |
0.214 |
-0.080 |
| 56 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.214 |
0.056 |
| 57 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
0.211 |
-0.034 |
| 58 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.204 |
-0.056 |
| 59 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.202 |
0.409 |
| 60 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.200 |
0.257 |
| 61 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.200 |
0.126 |
| 62 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.198 |
-0.071 |
| 63 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.197 |
0.051 |
| 64 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.197 |
-0.248 |
| 65 |
264761_at |
AT1G61280
|
[Phosphatidylinositol N-acetylglucosaminyltransferase, GPI19/PIG-P subunit] |
0.196 |
0.041 |
| 66 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.193 |
0.098 |
| 67 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.192 |
-0.000 |
| 68 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.181 |
-0.213 |
| 69 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.180 |
-0.057 |
| 70 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.180 |
-0.155 |
| 71 |
259979_at |
AT1G76600
|
unknown |
0.174 |
0.007 |
| 72 |
258935_at |
AT3G10120
|
unknown |
0.172 |
0.120 |
| 73 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.170 |
-0.078 |
| 74 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
0.168 |
0.192 |
| 75 |
251647_at |
AT3G57770
|
[Protein kinase superfamily protein] |
0.168 |
-0.112 |
| 76 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
0.168 |
-0.027 |
| 77 |
266275_at |
AT2G29370
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.166 |
-0.197 |
| 78 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.165 |
0.040 |
| 79 |
256603_at |
AT3G28270
|
unknown |
0.164 |
-0.496 |
| 80 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.163 |
0.003 |
| 81 |
258117_at |
AT3G14700
|
[SART-1 family] |
0.162 |
-0.108 |
| 82 |
246612_at |
AT5G35320
|
unknown |
0.161 |
0.016 |
| 83 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.161 |
-0.166 |
| 84 |
247287_at |
AT5G64230
|
unknown |
0.160 |
-0.006 |
| 85 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.159 |
-0.202 |
| 86 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.152 |
-0.311 |
| 87 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.152 |
-0.170 |
| 88 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.150 |
-0.028 |
| 89 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.149 |
0.039 |
| 90 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.147 |
0.043 |
| 91 |
264341_at |
AT1G70270
|
unknown |
0.147 |
-0.141 |
| 92 |
245313_at |
AT4G15420
|
[Ubiquitin fusion degradation UFD1 family protein] |
0.142 |
0.115 |
| 93 |
262514_at |
AT1G34190
|
anac017, NAC domain containing protein 17, NAC017, NAC domain containing protein 17 |
-0.798 |
-0.054 |
| 94 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.606 |
-0.038 |
| 95 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.406 |
-0.097 |
| 96 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.298 |
0.293 |
| 97 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
-0.282 |
0.081 |
| 98 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
-0.271 |
-0.196 |
| 99 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.259 |
-0.002 |
| 100 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.255 |
0.093 |
| 101 |
261221_at |
AT1G19960
|
unknown |
-0.245 |
0.019 |
| 102 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.230 |
0.042 |
| 103 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
-0.224 |
0.068 |
| 104 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.223 |
0.036 |
| 105 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.222 |
-0.186 |
| 106 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.220 |
0.051 |
| 107 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.217 |
-0.103 |
| 108 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.213 |
0.014 |
| 109 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.211 |
0.023 |
| 110 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.207 |
-0.121 |
| 111 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.203 |
-0.097 |
| 112 |
259276_at |
AT3G01190
|
[Peroxidase superfamily protein] |
-0.197 |
0.040 |
| 113 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.189 |
0.018 |
| 114 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.189 |
0.059 |
| 115 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.186 |
0.301 |
| 116 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.185 |
-0.011 |
| 117 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.182 |
0.018 |
| 118 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.180 |
0.026 |
| 119 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.172 |
0.223 |
| 120 |
249167_at |
AT5G42860
|
unknown |
-0.170 |
0.015 |
| 121 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.168 |
-0.108 |
| 122 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.167 |
-0.263 |
| 123 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.163 |
0.296 |
| 124 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-0.163 |
0.275 |
| 125 |
262317_at |
AT2G48140
|
EDA4, embryo sac development arrest 4 |
-0.154 |
-0.079 |
| 126 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
-0.151 |
-0.045 |
| 127 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.148 |
0.023 |
| 128 |
266167_at |
AT2G38860
|
DJ-1e, DJ1E, YLS5 |
-0.146 |
-0.161 |
| 129 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.142 |
-0.141 |
| 130 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
-0.142 |
0.044 |
| 131 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.141 |
-0.047 |
| 132 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.139 |
0.033 |
| 133 |
265611_at |
AT2G25510
|
unknown |
-0.136 |
0.007 |
| 134 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
-0.135 |
-0.149 |
| 135 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.134 |
0.122 |
| 136 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.131 |
-0.164 |
| 137 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.129 |
0.105 |
| 138 |
264680_at |
AT1G65510
|
unknown |
-0.127 |
0.022 |
| 139 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.124 |
-0.184 |
| 140 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.123 |
-0.148 |
| 141 |
252718_at |
AT3G43940
|
unknown |
-0.121 |
0.002 |
| 142 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.121 |
-0.173 |
| 143 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
-0.115 |
0.049 |
| 144 |
247059_at |
AT5G66690
|
UGT72E2 |
-0.115 |
-0.002 |
| 145 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
-0.113 |
-0.001 |
| 146 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.112 |
-0.052 |
| 147 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
-0.112 |
0.087 |
| 148 |
264998_at |
AT1G67330
|
unknown |
-0.110 |
-0.005 |
| 149 |
257924_at |
AT3G23190
|
[HR-like lesion-inducing protein-related] |
-0.110 |
0.023 |
| 150 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.109 |
0.139 |
| 151 |
262529_at |
AT1G17250
|
AtRLP3, receptor like protein 3, RFO2, RESISTANCE TO FUSARIUM OXYSPORUM 2, RLP3, receptor like protein 3 |
-0.108 |
0.154 |
| 152 |
248942_at |
AT5G45480
|
unknown |
-0.105 |
-0.124 |
| 153 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.105 |
-0.012 |
| 154 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.104 |
-0.267 |
| 155 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
-0.104 |
-0.005 |
| 156 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.102 |
-0.047 |
| 157 |
265188_at |
AT1G23800
|
ALDH2B, aldehyde dehydrogenase 2B, ALDH2B7, aldehyde dehydrogenase 2B7 |
-0.102 |
0.006 |
| 158 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
-0.102 |
-0.037 |
| 159 |
245127_at |
AT2G47600
|
ATMHX, magnesium/proton exchanger, ATMHX1, MHX, magnesium/proton exchanger, MHX1, MAGNESIUM/PROTON EXCHANGER 1 |
-0.101 |
0.110 |
| 160 |
252095_at |
AT3G51000
|
[alpha/beta-Hydrolases superfamily protein] |
-0.101 |
0.025 |
| 161 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.100 |
-0.122 |
| 162 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.100 |
0.030 |
| 163 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.100 |
-0.060 |
| 164 |
257305_at |
AT3G30250
|
unknown |
-0.099 |
-0.069 |
| 165 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
-0.099 |
0.018 |
| 166 |
262103_at |
AT1G02940
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, GSTF5, glutathione S-transferase (class phi) 5 |
-0.099 |
-0.137 |
| 167 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.099 |
-0.237 |
| 168 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.098 |
-0.505 |
| 169 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.098 |
0.008 |
| 170 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.097 |
0.049 |
| 171 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.097 |
0.026 |
| 172 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
-0.096 |
0.037 |
| 173 |
245596_at |
AT4G14180
|
AtPRD1, putative recombination initiation defect 1, PRD1, putative recombination initiation defect 1 |
-0.095 |
0.061 |
| 174 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
-0.095 |
0.160 |
| 175 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
-0.095 |
-0.055 |
| 176 |
249221_at |
AT5G42440
|
[Protein kinase superfamily protein] |
-0.095 |
0.031 |
| 177 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
-0.095 |
-0.093 |
| 178 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
-0.094 |
0.047 |
| 179 |
258047_at |
AT3G21240
|
4CL2, 4-coumarate:CoA ligase 2, AT4CL2 |
-0.093 |
0.059 |
| 180 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.093 |
-0.165 |
| 181 |
260254_at |
AT1G74210
|
AtGDPD5, GDPD5, glycerophosphodiester phosphodiesterase 5 |
-0.092 |
0.123 |
| 182 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
-0.092 |
-0.134 |
| 183 |
252519_at |
AT3G46360
|
unknown |
-0.092 |
0.104 |