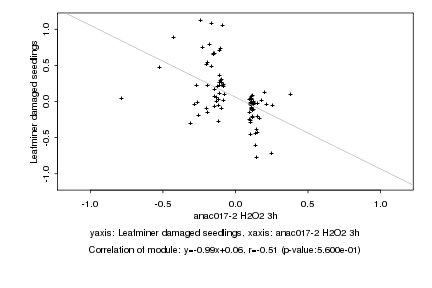
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
anac017-2 H2O2 3h |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
247224_at |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
0.379 |
0.099 |
| 2 |
263165_at |
AT1G03060
|
SPI, SPIRRIG |
0.255 |
-0.049 |
| 3 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
0.245 |
-0.712 |
| 4 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.214 |
-0.038 |
| 5 |
263515_at |
AT2G21640
|
unknown |
0.197 |
0.136 |
| 6 |
266618_at |
AT2G35480
|
unknown |
0.174 |
0.026 |
| 7 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.162 |
-0.229 |
| 8 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
0.148 |
-0.422 |
| 9 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
0.148 |
-0.206 |
| 10 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.146 |
-0.023 |
| 11 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
0.145 |
-0.767 |
| 12 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
0.145 |
-0.380 |
| 13 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
0.137 |
-0.604 |
| 14 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
0.133 |
-0.442 |
| 15 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.130 |
-0.020 |
| 16 |
265644_at |
AT2G27380
|
ATEPR1, extensin proline-rich 1, EPR1, extensin proline-rich 1 |
0.123 |
-0.041 |
| 17 |
250345_at |
AT5G11940
|
[Subtilase family protein] |
0.123 |
-0.001 |
| 18 |
256818_at |
AT3G21420
|
LBO1, LATERAL BRANCHING OXIDOREDUCTASE 1 |
0.120 |
-0.099 |
| 19 |
263525_at |
AT2G24840
|
AGL61, AGAMOUS-like 61, DIA, DIANA |
0.119 |
0.007 |
| 20 |
250653_at |
AT5G06930
|
[LOCATED IN: chloroplast] |
0.115 |
-0.120 |
| 21 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
0.114 |
-0.198 |
| 22 |
249574_at |
AT5G37660
|
PDLP7, plasmodesmata-located protein 7 |
0.112 |
-0.212 |
| 23 |
260163_at |
AT1G79900
|
ATMBAC2, RABIDOPSIS MITOCHONDRIAL BASIC AMINO ACID CARRIER 2, BAC2 |
0.112 |
0.093 |
| 24 |
266272_at |
AT2G29600
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.110 |
-0.054 |
| 25 |
265647_at |
AT2G27410
|
unknown |
0.110 |
-0.021 |
| 26 |
248672_at |
AT5G48770
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.109 |
-0.002 |
| 27 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
0.109 |
0.032 |
| 28 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.107 |
-0.034 |
| 29 |
246898_at |
AT5G25580
|
unknown |
0.107 |
-0.090 |
| 30 |
266470_at |
AT2G31080
|
[non-LTR retrotransposon family (LINE), has a 1.3e-49 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
0.107 |
-0.070 |
| 31 |
253352_at |
AT4G33710
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.106 |
-0.100 |
| 32 |
262764_at |
AT1G13140
|
CYP86C3, cytochrome P450, family 86, subfamily C, polypeptide 3 |
0.105 |
0.078 |
| 33 |
263769_at |
AT2G06390
|
transposable element gene |
0.105 |
-0.045 |
| 34 |
250808_at |
AT5G05150
|
ATATG18E, ARABIDOPSIS THALIANA HOMOLOG OF YEAST AUTOPHAGY 18E, ATG18E, autophagy-related gene 18E, G18E, autophagy-related gene 18E |
0.104 |
0.050 |
| 35 |
245224_at |
AT3G29796
|
unknown |
0.104 |
-0.005 |
| 36 |
249058_at |
AT5G44510
|
TAO1, target of AVRB operation1 |
0.102 |
-0.251 |
| 37 |
252205_at |
AT3G50350
|
unknown |
0.102 |
-0.025 |
| 38 |
264997_at |
AT1G67220
|
ATHPCAT1, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 1, HAC02, HAC2, histone acetyltransferase of the CBP family 2 |
0.100 |
-0.021 |
| 39 |
248080_at |
AT5G55380
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
0.100 |
-0.450 |
| 40 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.100 |
-0.281 |
| 41 |
258620_at |
AT3G02940
|
AtMYB107, myb domain protein 107, MYB107, myb domain protein 107 |
0.100 |
0.045 |
| 42 |
260905_at |
AT1G02710
|
[glycine-rich protein] |
0.098 |
-0.014 |
| 43 |
262031_x_at |
AT1G37160
|
[gypsy-like retrotransposon family (Athila), similar to putative Athila retroelement ORF1 protein GI:4567296 from (Arabidopsis thaliana)] |
0.097 |
-0.002 |
| 44 |
245858_at |
AT5G28280
|
[pseudogene] |
0.097 |
-0.034 |
| 45 |
250551_at |
AT5G07880
|
ATSNAP29, ARABIDOPSIS THALIANA SYNAPTOSOMAL-ASSOCIATED PROTEIN SNAP25-LIKE 29, SNAP29, synaptosomal-associated protein SNAP25-like 29 |
0.095 |
0.030 |
| 46 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
0.095 |
-0.141 |
| 47 |
251365_at |
AT3G61310
|
AHL11, AT-hook motif nuclear localized protein 11 |
0.094 |
-0.246 |
| 48 |
262514_at |
AT1G34190
|
anac017, NAC domain containing protein 17, NAC017, NAC domain containing protein 17 |
-0.789 |
0.053 |
| 49 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.526 |
0.474 |
| 50 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.427 |
0.894 |
| 51 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
-0.314 |
-0.294 |
| 52 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.289 |
-0.036 |
| 53 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
-0.270 |
0.230 |
| 54 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-0.262 |
-0.005 |
| 55 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
-0.258 |
-0.188 |
| 56 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
-0.246 |
1.135 |
| 57 |
256442_at |
AT3G10930
|
unknown |
-0.228 |
0.757 |
| 58 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.204 |
-0.091 |
| 59 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
-0.204 |
0.517 |
| 60 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.197 |
0.235 |
| 61 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.197 |
0.543 |
| 62 |
259222_at |
AT3G03680
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.195 |
-0.144 |
| 63 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.180 |
0.803 |
| 64 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.170 |
1.092 |
| 65 |
252470_at |
AT3G46930
|
[Protein kinase superfamily protein] |
-0.169 |
0.496 |
| 66 |
250158_at |
AT5G15190
|
unknown |
-0.158 |
0.656 |
| 67 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
-0.157 |
0.672 |
| 68 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
-0.151 |
0.072 |
| 69 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.151 |
-0.062 |
| 70 |
254390_at |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
-0.150 |
0.171 |
| 71 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.149 |
0.671 |
| 72 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
-0.137 |
0.064 |
| 73 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.135 |
0.004 |
| 74 |
255900_at |
AT1G17830
|
unknown |
-0.128 |
0.208 |
| 75 |
247800_at |
AT5G58570
|
unknown |
-0.123 |
-0.044 |
| 76 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.122 |
0.035 |
| 77 |
261817_at |
AT1G08180
|
unknown |
-0.117 |
-0.271 |
| 78 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-0.116 |
0.373 |
| 79 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.114 |
0.267 |
| 80 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
-0.113 |
0.718 |
| 81 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
-0.111 |
0.116 |
| 82 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.110 |
0.231 |
| 83 |
260627_at |
AT1G62310
|
[transcription factor jumonji (jmjC) domain-containing protein] |
-0.109 |
0.301 |
| 84 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
-0.109 |
0.741 |
| 85 |
260439_at |
AT1G68340
|
unknown |
-0.102 |
0.277 |
| 86 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.100 |
0.306 |
| 87 |
258702_at |
AT3G09730
|
unknown |
-0.098 |
-0.093 |
| 88 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
-0.096 |
0.229 |
| 89 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.092 |
1.058 |
| 90 |
245410_at |
AT4G17220
|
ATMAP70-5, microtubule-associated proteins 70-5, MAP70-5, microtubule-associated proteins 70-5 |
-0.088 |
0.016 |
| 91 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
-0.088 |
0.239 |
| 92 |
251114_at |
AT5G01380
|
[Homeodomain-like superfamily protein] |
-0.087 |
0.213 |
| 93 |
254060_at |
AT4G25350
|
SHB1, SHORT HYPOCOTYL UNDER BLUE1 |
-0.082 |
0.102 |