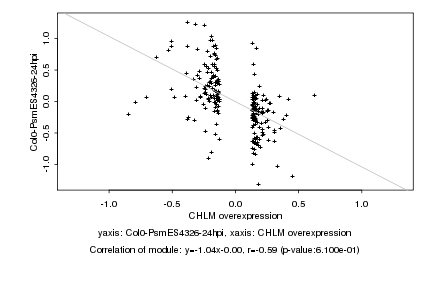
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
CHLM overexpression |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
258289_at |
AT3G23450
|
unknown |
0.624 |
0.101 |
| 2 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.450 |
-1.178 |
| 3 |
263285_at |
AT2G36120
|
DOT1, DEFECTIVELY ORGANIZED TRIBUTARIES 1 |
0.418 |
0.041 |
| 4 |
245002_at |
ATCG00270
|
PSBD, photosystem II reaction center protein D |
0.403 |
-0.212 |
| 5 |
244934_at |
ATCG01080
|
NDHG |
0.378 |
-0.281 |
| 6 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.349 |
-0.426 |
| 7 |
248862_at |
AT5G46730
|
[glycine-rich protein] |
0.346 |
0.081 |
| 8 |
257057_at |
AT3G15310
|
unknown |
0.327 |
-1.019 |
| 9 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.305 |
-0.491 |
| 10 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.304 |
-0.455 |
| 11 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.303 |
-0.634 |
| 12 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.298 |
-0.162 |
| 13 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
0.275 |
-0.023 |
| 14 |
265847_at |
AT2G35750
|
unknown |
0.262 |
-0.027 |
| 15 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
0.260 |
-0.400 |
| 16 |
254105_at |
AT4G25080
|
CHLM, magnesium-protoporphyrin IX methyltransferase |
0.258 |
-0.610 |
| 17 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.255 |
-0.141 |
| 18 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.252 |
-0.135 |
| 19 |
264636_at |
AT1G65490
|
unknown |
0.247 |
-0.299 |
| 20 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.247 |
-0.157 |
| 21 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.241 |
0.038 |
| 22 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
0.234 |
-0.328 |
| 23 |
245486_x_at |
AT4G16240
|
unknown |
0.232 |
0.019 |
| 24 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.220 |
-0.512 |
| 25 |
258220_at |
AT3G17830
|
DJA4, DNA J protein A4 |
0.219 |
-0.172 |
| 26 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
0.216 |
0.102 |
| 27 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.213 |
0.022 |
| 28 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
0.213 |
-0.439 |
| 29 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.212 |
-0.109 |
| 30 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.212 |
-0.478 |
| 31 |
266384_at |
AT2G14660
|
unknown |
0.211 |
-0.532 |
| 32 |
265831_at |
AT2G14460
|
unknown |
0.210 |
-0.176 |
| 33 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
0.207 |
-0.166 |
| 34 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.204 |
-0.345 |
| 35 |
254208_at |
AT4G24175
|
unknown |
0.201 |
-0.162 |
| 36 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
0.195 |
-0.715 |
| 37 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.188 |
-0.103 |
| 38 |
254586_at |
AT4G19510
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.184 |
0.253 |
| 39 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.184 |
-0.411 |
| 40 |
263840_at |
AT2G36885
|
unknown |
0.183 |
-0.592 |
| 41 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.181 |
-1.303 |
| 42 |
261876_at |
AT1G50590
|
[RmlC-like cupins superfamily protein] |
0.180 |
-0.150 |
| 43 |
260676_at |
AT1G19450
|
[Major facilitator superfamily protein] |
0.177 |
-0.196 |
| 44 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.176 |
-0.686 |
| 45 |
250696_at |
AT5G06790
|
unknown |
0.172 |
-0.567 |
| 46 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
0.171 |
0.126 |
| 47 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
0.170 |
-0.033 |
| 48 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
0.169 |
-0.328 |
| 49 |
254145_at |
AT4G24700
|
unknown |
0.168 |
-0.641 |
| 50 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.165 |
0.852 |
| 51 |
245426_at |
AT4G17540
|
unknown |
0.164 |
-0.570 |
| 52 |
264761_at |
AT1G61280
|
[Phosphatidylinositol N-acetylglucosaminyltransferase, GPI19/PIG-P subunit] |
0.161 |
0.060 |
| 53 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
0.160 |
-0.114 |
| 54 |
261350_at |
AT1G79770
|
unknown |
0.159 |
-0.178 |
| 55 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.159 |
-0.676 |
| 56 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
0.157 |
-0.359 |
| 57 |
261406_at |
AT1G18800
|
NRP2, NAP1-related protein 2 |
0.156 |
-0.310 |
| 58 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
0.155 |
-0.258 |
| 59 |
256653_at |
AT3G18870
|
[Mitochondrial transcription termination factor family protein] |
0.155 |
-0.278 |
| 60 |
245743_at |
AT1G51080
|
unknown |
0.154 |
-0.647 |
| 61 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.154 |
-0.826 |
| 62 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
0.154 |
-0.603 |
| 63 |
255777_at |
AT1G18630
|
GR-RBP6, glycine-rich RNA-binding protein 6 |
0.153 |
0.106 |
| 64 |
252957_at |
AT4G38680
|
ATCSP2, ARABIDOPSIS THALIANA COLD SHOCK PROTEIN 2, CSDP2, COLD SHOCK DOMAIN PROTEIN 2, CSP2, COLD SHOCK PROTEIN 2, GRP2, glycine rich protein 2 |
0.153 |
-0.287 |
| 65 |
249357_at |
AT5G40490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.153 |
0.050 |
| 66 |
249470_at |
AT5G39350
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.152 |
-0.073 |
| 67 |
264087_at |
AT2G31340
|
emb1381, embryo defective 1381 |
0.152 |
-0.133 |
| 68 |
249676_at |
AT5G35960
|
[Protein kinase family protein] |
0.151 |
0.033 |
| 69 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
0.151 |
0.060 |
| 70 |
266002_at |
AT2G37310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.150 |
0.070 |
| 71 |
260100_at |
AT1G73177
|
APC13, anaphase-promoting complex 13, BNS, BONSAI |
0.150 |
-0.096 |
| 72 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.149 |
-0.228 |
| 73 |
250722_at |
AT5G06190
|
unknown |
0.148 |
0.445 |
| 74 |
252661_at |
AT3G44450
|
unknown |
0.148 |
-0.080 |
| 75 |
248940_at |
AT5G45400
|
ATRPA70C, RPA70C |
0.147 |
-0.029 |
| 76 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.146 |
-0.134 |
| 77 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.146 |
0.159 |
| 78 |
259696_at |
AT1G63150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.146 |
-0.007 |
| 79 |
251730_at |
AT3G56330
|
[N2,N2-dimethylguanosine tRNA methyltransferase] |
0.146 |
-0.293 |
| 80 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.146 |
-0.750 |
| 81 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.144 |
-0.244 |
| 82 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.144 |
-0.504 |
| 83 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
0.144 |
-0.586 |
| 84 |
262028_at |
AT1G35560
|
AtTCP23, TCP23, TCP domain protein 23 |
0.143 |
-0.140 |
| 85 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
0.143 |
-0.655 |
| 86 |
248954_at |
AT5G45420
|
maMYB, membrane anchored MYB |
0.142 |
0.046 |
| 87 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.142 |
0.109 |
| 88 |
252654_at |
AT3G44740
|
[Class II aaRS and biotin synthetases superfamily protein] |
0.142 |
-0.227 |
| 89 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.142 |
0.594 |
| 90 |
251727_at |
AT3G56290
|
unknown |
0.141 |
-0.132 |
| 91 |
257968_at |
AT3G27550
|
[RNA-binding CRS1 / YhbY (CRM) domain protein] |
0.141 |
-0.065 |
| 92 |
248136_at |
AT5G54910
|
[DEA(D/H)-box RNA helicase family protein] |
0.141 |
-0.286 |
| 93 |
252410_at |
AT3G47450
|
ATNOA1, ATNOS1, NOA1, NO ASSOCIATED 1, NOS1, NITRIC OXIDE SYNTHASE 1, RIF1, RESISTANT TO INHIBITION WITH FOSMIDOMYCIN 1 |
0.141 |
-0.376 |
| 94 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.141 |
-0.180 |
| 95 |
245518_at |
AT4G15850
|
ATRH1, RNA helicase 1, RH1, RNA helicase 1 |
0.140 |
-0.251 |
| 96 |
263471_at |
AT2G31890
|
ATRAP, RAP |
0.139 |
-0.372 |
| 97 |
256837_at |
AT3G22900
|
NRPD7 |
0.138 |
-0.207 |
| 98 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
0.138 |
-0.628 |
| 99 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
0.138 |
-0.136 |
| 100 |
250421_at |
AT5G11270
|
OCP3, overexpressor of cationic peroxidase 3 |
0.138 |
-0.202 |
| 101 |
244998_at |
ATCG00180
|
RPOC1 |
0.138 |
-0.239 |
| 102 |
263674_at |
AT2G04790
|
unknown |
0.136 |
-0.815 |
| 103 |
252508_at |
AT3G46210
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.136 |
-0.299 |
| 104 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
0.136 |
-0.007 |
| 105 |
249425_at |
AT5G39790
|
5'-AMP-activated protein kinase-related |
0.136 |
-0.146 |
| 106 |
258701_at |
AT3G09720
|
AtRH57, RH57, RNA helicase 57 |
0.135 |
-0.200 |
| 107 |
261790_at |
AT1G16000
|
unknown |
0.135 |
-0.104 |
| 108 |
264002_at |
AT2G22360
|
DJA6, DNA J protein A6 |
0.134 |
-0.046 |
| 109 |
258082_at |
AT3G25905
|
CLE27, CLAVATA3/ESR-RELATED 27 |
0.134 |
-0.130 |
| 110 |
245049_at |
ATCG00050
|
RPS16, ribosomal protein S16 |
0.134 |
-0.731 |
| 111 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
0.133 |
-0.254 |
| 112 |
257910_at |
AT3G25580
|
[Thioredoxin superfamily protein] |
0.132 |
0.035 |
| 113 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
0.132 |
-0.634 |
| 114 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.132 |
0.926 |
| 115 |
261772_at |
AT1G76240
|
unknown |
0.132 |
0.075 |
| 116 |
258424_at |
AT3G16750
|
unknown |
0.132 |
-0.398 |
| 117 |
256985_at |
AT3G13540
|
ATMYB5, myb domain protein 5, MYB5, myb domain protein 5 |
0.131 |
0.129 |
| 118 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.131 |
-0.247 |
| 119 |
261003_at |
AT1G26500
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.131 |
-0.048 |
| 120 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.130 |
-0.132 |
| 121 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.130 |
-0.219 |
| 122 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.130 |
-0.989 |
| 123 |
248079_at |
AT5G55790
|
unknown |
0.129 |
-0.112 |
| 124 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.853 |
-0.198 |
| 125 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-0.793 |
-0.006 |
| 126 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.710 |
0.065 |
| 127 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.631 |
0.712 |
| 128 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.533 |
0.813 |
| 129 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.513 |
0.205 |
| 130 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.513 |
0.883 |
| 131 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.507 |
0.969 |
| 132 |
256631_at |
AT3G28320
|
unknown |
-0.486 |
0.067 |
| 133 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.402 |
0.087 |
| 134 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.395 |
0.450 |
| 135 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.384 |
0.876 |
| 136 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.384 |
-0.272 |
| 137 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.383 |
1.271 |
| 138 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.372 |
-0.242 |
| 139 |
265539_at |
AT2G15830
|
unknown |
-0.338 |
0.365 |
| 140 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.327 |
-0.297 |
| 141 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
-0.318 |
1.236 |
| 142 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.314 |
0.031 |
| 143 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.313 |
0.224 |
| 144 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.305 |
0.841 |
| 145 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.305 |
0.417 |
| 146 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.290 |
0.373 |
| 147 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.285 |
0.487 |
| 148 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.282 |
0.075 |
| 149 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.282 |
0.091 |
| 150 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.253 |
-0.036 |
| 151 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
-0.249 |
1.223 |
| 152 |
264456_at |
AT1G10390
|
[Nucleoporin autopeptidase] |
-0.248 |
0.211 |
| 153 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.247 |
0.141 |
| 154 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.246 |
0.597 |
| 155 |
259766_at |
AT1G64360
|
unknown |
-0.244 |
0.248 |
| 156 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-0.242 |
-0.105 |
| 157 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
-0.241 |
0.193 |
| 158 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.239 |
-0.461 |
| 159 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.238 |
0.154 |
| 160 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.235 |
0.562 |
| 161 |
254894_at |
AT4G11840
|
PLDGAMMA3, phospholipase D gamma 3 |
-0.232 |
0.572 |
| 162 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.230 |
0.120 |
| 163 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.226 |
0.474 |
| 164 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
-0.224 |
0.802 |
| 165 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.222 |
0.265 |
| 166 |
245692_at |
AT5G04150
|
BHLH101 |
-0.221 |
0.062 |
| 167 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.221 |
0.540 |
| 168 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-0.220 |
0.046 |
| 169 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.215 |
-0.895 |
| 170 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.213 |
0.307 |
| 171 |
257621_at |
AT3G20410
|
CPK9, calmodulin-domain protein kinase 9 |
-0.213 |
0.254 |
| 172 |
261387_at |
AT1G05410
|
unknown |
-0.209 |
0.013 |
| 173 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
-0.205 |
0.107 |
| 174 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.204 |
0.720 |
| 175 |
258225_at |
AT3G15630
|
unknown |
-0.198 |
0.325 |
| 176 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.198 |
0.984 |
| 177 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.197 |
0.068 |
| 178 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.195 |
0.373 |
| 179 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.194 |
1.041 |
| 180 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.194 |
0.299 |
| 181 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
-0.193 |
0.464 |
| 182 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.193 |
-0.808 |
| 183 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
-0.188 |
0.975 |
| 184 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.187 |
0.210 |
| 185 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.183 |
0.603 |
| 186 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.181 |
0.104 |
| 187 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
-0.180 |
0.424 |
| 188 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
-0.180 |
0.382 |
| 189 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.178 |
0.404 |
| 190 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.175 |
0.883 |
| 191 |
253550_at |
AT4G30960
|
ATCIPK6, CBL-INTERACTING PROTEIN KINASE 6, CIPK6, CBL-INTERACTING PROTEIN KINASE 6, SIP3, SOS3-interacting protein 3, SNRK3.14, SNF1-RELATED PROTEIN KINASE 3.14 |
-0.173 |
-0.004 |
| 192 |
248294_at |
AT5G53060
|
RCF3, regulator of CBF gene expression 3, SHI1, SHINY 1 |
-0.170 |
0.187 |
| 193 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.169 |
0.776 |
| 194 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
-0.169 |
0.763 |
| 195 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.168 |
-0.150 |
| 196 |
265025_at |
AT1G24575
|
unknown |
-0.168 |
0.564 |
| 197 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.168 |
0.560 |
| 198 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.167 |
0.059 |
| 199 |
251012_at |
AT5G02580
|
unknown |
-0.166 |
0.592 |
| 200 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.165 |
0.400 |
| 201 |
253865_at |
AT4G27470
|
ATRMA3, RMA3, RING membrane-anchor 3 |
-0.164 |
0.257 |
| 202 |
260179_at |
AT1G70690
|
HWI1, HOPW1-1-INDUCED GENE1, PDLP5, PLASMODESMATA-LOCATED PROTEIN 5 |
-0.164 |
0.733 |
| 203 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.163 |
0.407 |
| 204 |
246666_at |
AT5G34830
|
unknown |
-0.162 |
-0.001 |
| 205 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
-0.162 |
0.898 |
| 206 |
264828_at |
AT1G03380
|
ATATG18G, homolog of yeast autophagy 18 (ATG18) G, ATG18G, homolog of yeast autophagy 18 (ATG18) G |
-0.162 |
0.162 |
| 207 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.159 |
-0.513 |
| 208 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.159 |
0.308 |
| 209 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
-0.158 |
-0.084 |
| 210 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
-0.157 |
0.525 |
| 211 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.156 |
0.307 |
| 212 |
264548_at |
AT1G55680
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.155 |
0.049 |
| 213 |
245398_at |
AT4G14900
|
[FRIGIDA-like protein] |
-0.153 |
0.308 |
| 214 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
-0.152 |
0.669 |
| 215 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.152 |
-0.361 |
| 216 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
-0.151 |
0.854 |
| 217 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.151 |
-0.042 |
| 218 |
266355_at |
AT2G01400
|
unknown |
-0.151 |
0.095 |
| 219 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.150 |
0.171 |
| 220 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.149 |
0.120 |
| 221 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.149 |
-0.135 |
| 222 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
-0.148 |
0.697 |
| 223 |
261659_at |
AT1G50030
|
TOR, target of rapamycin |
-0.148 |
0.084 |
| 224 |
251468_at |
AT3G59290
|
[ENTH/VHS family protein] |
-0.147 |
0.336 |
| 225 |
258204_at |
AT3G13960
|
AtGRF5, growth-regulating factor 5, GRF5, growth-regulating factor 5 |
-0.146 |
0.063 |
| 226 |
263183_at |
AT1G05570
|
ATGSL06, ATGSL6, CALS1, callose synthase 1, GSL06, GSL6, GLUCAN SYNTHASE-LIKE 6 |
-0.143 |
0.297 |
| 227 |
258939_at |
AT3G10020
|
unknown |
-0.142 |
0.025 |
| 228 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
-0.142 |
-0.150 |
| 229 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.142 |
0.498 |
| 230 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.142 |
0.330 |
| 231 |
257883_at |
AT3G16940
|
[calmodulin binding] |
-0.141 |
0.141 |
| 232 |
261711_at |
AT1G32700
|
[PLATZ transcription factor family protein] |
-0.141 |
0.144 |
| 233 |
251051_at |
AT5G02460
|
[Dof-type zinc finger DNA-binding family protein] |
-0.140 |
0.069 |
| 234 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
-0.139 |
-0.189 |
| 235 |
257174_at |
AT3G27190
|
UKL2, uridine kinase-like 2 |
-0.139 |
0.034 |
| 236 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
-0.139 |
0.329 |
| 237 |
257216_at |
AT3G14990
|
AtDJ1A, DJ-1 homolog A, DJ-1a, DJ1A, DJ-1 homolog A |
-0.139 |
0.331 |
| 238 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.136 |
0.170 |
| 239 |
245873_at |
AT1G26260
|
CIB5, cryptochrome-interacting basic-helix-loop-helix 5 |
-0.136 |
-0.077 |
| 240 |
259312_at |
AT3G05200
|
ATL6, Arabidopsis toxicos en levadura 6 |
-0.136 |
0.362 |
| 241 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.132 |
0.044 |
| 242 |
253808_at |
AT4G28300
|
unknown |
-0.130 |
0.274 |
| 243 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
-0.129 |
0.043 |
| 244 |
258455_at |
AT3G22440
|
[FRIGIDA-like protein] |
-0.129 |
0.008 |
| 245 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.128 |
-0.600 |