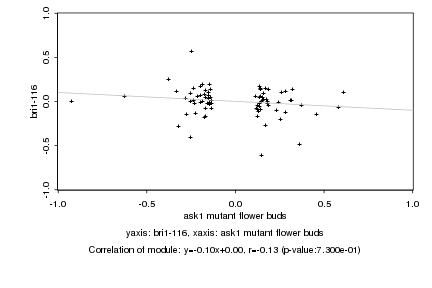
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ask1 mutant flower buds |
Log2 signal ratio
bri1-116 |
| 1 |
244994_at |
ATCG01010
|
NDHF |
0.604 |
0.104 |
| 2 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.579 |
-0.067 |
| 3 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
0.455 |
-0.141 |
| 4 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.369 |
-0.039 |
| 5 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.358 |
-0.484 |
| 6 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.317 |
0.146 |
| 7 |
262211_at |
AT1G74930
|
ORA47 |
0.315 |
0.018 |
| 8 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.305 |
0.021 |
| 9 |
262135_at |
AT1G78080
|
AtWIND1, RAP2.4, related to AP2 4, WIND1, wound induced dedifferentiation 1 |
0.282 |
0.120 |
| 10 |
267623_at |
AT2G39650
|
unknown |
0.282 |
-0.116 |
| 11 |
247177_at |
AT5G65300
|
unknown |
0.256 |
0.112 |
| 12 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
0.249 |
-0.196 |
| 13 |
252474_at |
AT3G46620
|
unknown |
0.240 |
-0.003 |
| 14 |
254158_at |
AT4G24380
|
[INVOLVED IN: 10-formyltetrahydrofolate biosynthetic process, folic acid and derivative biosynthetic process] |
0.231 |
-0.096 |
| 15 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.184 |
-0.045 |
| 16 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.182 |
0.141 |
| 17 |
253833_at |
AT4G27790
|
[Calcium-binding EF hand family protein] |
0.180 |
0.005 |
| 18 |
259526_at |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.179 |
-0.012 |
| 19 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.173 |
0.027 |
| 20 |
250432_at |
AT5G10420
|
[MATE efflux family protein] |
0.167 |
0.149 |
| 21 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.164 |
-0.269 |
| 22 |
248373_at |
AT5G51860
|
AGL72, AGAMOUS-like 72 |
0.158 |
0.027 |
| 23 |
260155_at |
AT1G52870
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.155 |
0.102 |
| 24 |
244936_at |
ATCG01100
|
NDHA |
0.152 |
0.020 |
| 25 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
0.148 |
0.052 |
| 26 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.147 |
-0.610 |
| 27 |
258019_at |
AT3G19470
|
[F-box and associated interaction domains-containing protein] |
0.145 |
0.002 |
| 28 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
0.141 |
-0.084 |
| 29 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.137 |
0.058 |
| 30 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.136 |
0.138 |
| 31 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.136 |
0.156 |
| 32 |
262605_at |
AT1G15170
|
[MATE efflux family protein] |
0.135 |
0.053 |
| 33 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.133 |
0.177 |
| 34 |
247861_at |
AT5G58160
|
[actin binding] |
0.132 |
-0.012 |
| 35 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.132 |
-0.049 |
| 36 |
266424_at |
AT2G41330
|
[Glutaredoxin family protein] |
0.128 |
-0.108 |
| 37 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
0.124 |
-0.169 |
| 38 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.119 |
-0.084 |
| 39 |
245003_at |
ATCG00280
|
PSBC, photosystem II reaction center protein C |
0.119 |
-0.036 |
| 40 |
250734_at |
AT5G06270
|
unknown |
0.115 |
-0.073 |
| 41 |
261895_at |
AT1G80830
|
ATNRAMP1, NRAMP1, natural resistance-associated macrophage protein 1, PMIT1 |
0.111 |
0.059 |
| 42 |
262676_at |
AT1G75950
|
ASK1, ARABIDOPSIS SKP1 HOMOLOGUE 1, ATSKP1, SKP1, S phase kinase-associated protein 1, SKP1A, UIP1, UFO INTERACTING PROTEIN 1 |
-0.929 |
0.004 |
| 43 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.630 |
0.066 |
| 44 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
-0.382 |
0.259 |
| 45 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.337 |
0.124 |
| 46 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.323 |
-0.278 |
| 47 |
265552_at |
AT2G07560
|
AHA6, H(+)-ATPase 6, HA6, H(+)-ATPase 6 |
-0.282 |
0.041 |
| 48 |
260112_at |
AT1G63310
|
unknown |
-0.280 |
-0.142 |
| 49 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
-0.258 |
0.094 |
| 50 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.255 |
0.007 |
| 51 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.254 |
-0.398 |
| 52 |
257191_at |
AT3G13175
|
unknown |
-0.249 |
0.577 |
| 53 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
-0.238 |
0.148 |
| 54 |
247490_at |
AT5G61850
|
LFY, LEAFY, LFY3, LEAFY 3 |
-0.238 |
0.021 |
| 55 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
-0.234 |
-0.018 |
| 56 |
264288_at |
AT1G62045
|
unknown |
-0.227 |
-0.125 |
| 57 |
256584_at |
AT3G28750
|
unknown |
-0.218 |
0.066 |
| 58 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.200 |
0.173 |
| 59 |
266049_at |
AT2G40780
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.199 |
0.074 |
| 60 |
253540_at |
AT4G31615
|
[Transcriptional factor B3 family protein] |
-0.199 |
-0.008 |
| 61 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
-0.191 |
0.193 |
| 62 |
257105_at |
AT3G15300
|
[VQ motif-containing protein] |
-0.190 |
0.002 |
| 63 |
262843_at |
AT1G14687
|
AtHB32, homeobox protein 32, HB32, homeobox protein 32, ZHD14, ZINC FINGER HOMEODOMAIN 14 |
-0.178 |
0.081 |
| 64 |
262278_at |
AT1G68640
|
PAN, PERIANTHIA, TGA8, TGACG sequence-specific binding protein 8 |
-0.175 |
-0.181 |
| 65 |
254203_at |
AT4G24150
|
AtGRF8, growth-regulating factor 8, GRF8, growth-regulating factor 8 |
-0.173 |
0.047 |
| 66 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
-0.172 |
-0.163 |
| 67 |
254274_at |
AT4G22770
|
AHL2, AT-hook motif nuclear localized protein 2 |
-0.171 |
-0.078 |
| 68 |
245226_at |
AT3G29970
|
[B12D protein] |
-0.169 |
0.125 |
| 69 |
250636_at |
AT5G07520
|
ATGRP-8, ATGRP18, GRP18, glycine-rich protein 18 |
-0.163 |
-0.015 |
| 70 |
250097_at |
AT5G17280
|
unknown |
-0.155 |
0.045 |
| 71 |
244925_at |
ATMG00510
|
NAD7, NADH dehydrogenase subunit 7 |
-0.155 |
0.112 |
| 72 |
250174_at |
AT5G14380
|
AGP6, arabinogalactan protein 6 |
-0.153 |
0.079 |
| 73 |
265429_at |
AT2G20710
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.150 |
-0.010 |
| 74 |
265373_at |
AT2G06510
|
ATRPA1A, ARABIDOPSIS THALIANA REPLICATION PROTEIN A 1A, ATRPA70A, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT A, RPA1A, replication protein A 1A, RPA70A, RPA70-KDA SUBUNIT A |
-0.149 |
-0.016 |
| 75 |
257441_at |
AT2G04020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.147 |
-0.025 |
| 76 |
256942_at |
AT3G23290
|
LSH4, LIGHT SENSITIVE HYPOCOTYLS 4 |
-0.147 |
0.200 |
| 77 |
261311_at |
AT1G05770
|
[Mannose-binding lectin superfamily protein] |
-0.145 |
0.049 |
| 78 |
250708_at |
AT5G06070
|
RAB, RBE, RABBIT EARS |
-0.141 |
0.142 |
| 79 |
264621_at |
AT2G17700
|
STY8, serine/threonine/tyrosine kinase 8 |
-0.140 |
-0.074 |
| 80 |
260605_at |
AT2G43780
|
unknown |
-0.140 |
0.014 |
| 81 |
265849_at |
AT2G35736
|
unknown |
-0.139 |
-0.019 |