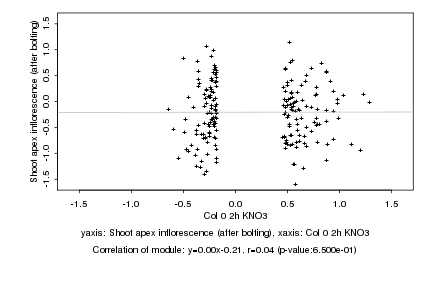
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col 0 2h KNO3 |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
255669_at |
AT4G00416
|
MBD3, methyl-CPG-binding domain 3 |
1.281 |
-0.007 |
| 2 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
1.225 |
0.148 |
| 3 |
250472_at |
AT5G10210
|
unknown |
1.201 |
-0.944 |
| 4 |
260624_at |
AT1G08100
|
ACH2, ATNRT2.2, NITRATE TRANSPORTER 2.2, NRT2.2, nitrate transporter 2.2, NRT2;2AT |
1.115 |
-0.822 |
| 5 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
1.031 |
0.120 |
| 6 |
259015_at |
AT3G07350
|
unknown |
0.983 |
-0.313 |
| 7 |
246142_at |
AT5G19970
|
unknown |
0.979 |
-0.031 |
| 8 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
0.974 |
0.053 |
| 9 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
0.942 |
0.212 |
| 10 |
253408_at |
AT4G32950
|
[Protein phosphatase 2C family protein] |
0.941 |
-0.725 |
| 11 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
0.935 |
-0.175 |
| 12 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.905 |
0.394 |
| 13 |
251301_at |
AT3G61880
|
CYP78A9, cytochrome p450 78a9 |
0.877 |
-0.820 |
| 14 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.873 |
0.598 |
| 15 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
0.872 |
-1.127 |
| 16 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
0.872 |
-0.159 |
| 17 |
252501_at |
AT3G46880
|
unknown |
0.871 |
-0.372 |
| 18 |
262399_at |
AT1G49500
|
unknown |
0.869 |
0.569 |
| 19 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
0.825 |
0.735 |
| 20 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.801 |
-0.433 |
| 21 |
266222_at |
AT2G28780
|
unknown |
0.785 |
-0.431 |
| 22 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
0.782 |
-0.150 |
| 23 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.781 |
-0.780 |
| 24 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
0.774 |
0.154 |
| 25 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.773 |
-0.319 |
| 26 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.773 |
-0.454 |
| 27 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.772 |
0.272 |
| 28 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.765 |
0.125 |
| 29 |
259996_at |
AT1G67910
|
unknown |
0.760 |
-0.389 |
| 30 |
255934_at |
AT1G12740
|
CYP87A2, cytochrome P450, family 87, subfamily A, polypeptide 2 |
0.730 |
-0.571 |
| 31 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
0.727 |
-0.108 |
| 32 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.722 |
0.642 |
| 33 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.683 |
-0.863 |
| 34 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
0.680 |
-0.085 |
| 35 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.679 |
0.518 |
| 36 |
265048_at |
AT1G52050
|
[Mannose-binding lectin superfamily protein] |
0.674 |
-0.486 |
| 37 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.668 |
0.402 |
| 38 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.657 |
-0.674 |
| 39 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
0.656 |
-0.804 |
| 40 |
260771_at |
AT1G49160
|
WNK7 |
0.654 |
-1.290 |
| 41 |
265049_at |
AT1G52060
|
[Mannose-binding lectin superfamily protein] |
0.637 |
0.024 |
| 42 |
247754_at |
AT5G59080
|
unknown |
0.635 |
-0.322 |
| 43 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
0.631 |
0.312 |
| 44 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.615 |
-0.653 |
| 45 |
247591_at |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
0.612 |
-0.158 |
| 46 |
257720_at |
AT3G18450
|
[PLAC8 family protein] |
0.608 |
-0.763 |
| 47 |
260754_at |
AT1G49000
|
unknown |
0.604 |
-0.139 |
| 48 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.595 |
-0.557 |
| 49 |
246054_at |
AT5G08360
|
unknown |
0.594 |
-0.436 |
| 50 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
0.588 |
0.188 |
| 51 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.583 |
-0.869 |
| 52 |
253317_at |
AT4G33960
|
unknown |
0.583 |
-0.341 |
| 53 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.583 |
0.011 |
| 54 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
0.581 |
-0.785 |
| 55 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.574 |
-1.583 |
| 56 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.571 |
-0.175 |
| 57 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.567 |
-0.009 |
| 58 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
0.562 |
-1.211 |
| 59 |
255377_at |
AT4G03500
|
[Ankyrin repeat family protein] |
0.557 |
-0.035 |
| 60 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
0.552 |
-1.204 |
| 61 |
255041_at |
AT4G09620
|
[Mitochondrial transcription termination factor family protein] |
0.550 |
-0.078 |
| 62 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.547 |
0.800 |
| 63 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.544 |
-0.819 |
| 64 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
0.542 |
0.087 |
| 65 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
0.539 |
-0.165 |
| 66 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.537 |
0.414 |
| 67 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.535 |
0.188 |
| 68 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
0.534 |
-0.142 |
| 69 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
0.534 |
-0.108 |
| 70 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.532 |
0.158 |
| 71 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
0.532 |
-0.650 |
| 72 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
0.526 |
-0.654 |
| 73 |
261481_at |
AT1G14260
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.525 |
0.770 |
| 74 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.522 |
0.067 |
| 75 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.521 |
-0.836 |
| 76 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.520 |
-0.040 |
| 77 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.518 |
-0.471 |
| 78 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.515 |
-0.436 |
| 79 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.512 |
1.146 |
| 80 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
0.506 |
0.054 |
| 81 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.505 |
-0.263 |
| 82 |
246238_at |
AT4G36670
|
AtPLT6, AtPMT6, PLT6, polyol transporter 6, PMT6, polyol/monosaccharide transporter 6 |
0.499 |
0.311 |
| 83 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.499 |
-0.827 |
| 84 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
0.496 |
-0.307 |
| 85 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
0.491 |
0.370 |
| 86 |
255924_at |
AT1G22170
|
[Phosphoglycerate mutase family protein] |
0.491 |
-0.068 |
| 87 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
0.487 |
-0.744 |
| 88 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
0.485 |
-0.786 |
| 89 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.484 |
0.006 |
| 90 |
246981_at |
AT5G04840
|
[bZIP protein] |
0.479 |
-0.200 |
| 91 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
0.479 |
-0.896 |
| 92 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.477 |
0.202 |
| 93 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.474 |
0.639 |
| 94 |
263465_at |
AT2G31940
|
unknown |
0.473 |
-0.108 |
| 95 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.472 |
0.631 |
| 96 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.472 |
-0.794 |
| 97 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
0.471 |
0.049 |
| 98 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
0.466 |
-0.664 |
| 99 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.464 |
-0.238 |
| 100 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
0.463 |
-0.688 |
| 101 |
266259_at |
AT2G27830
|
unknown |
0.458 |
-0.063 |
| 102 |
262459_at |
AT1G50400
|
[Eukaryotic porin family protein] |
0.454 |
0.272 |
| 103 |
246387_at |
AT1G77400
|
unknown |
0.452 |
-0.677 |
| 104 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.652 |
-0.138 |
| 105 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.596 |
-0.539 |
| 106 |
246333_at |
AT3G44840
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.551 |
-1.099 |
| 107 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.508 |
0.844 |
| 108 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.497 |
-0.587 |
| 109 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
-0.487 |
-0.340 |
| 110 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
-0.478 |
-0.925 |
| 111 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.454 |
-0.956 |
| 112 |
265063_at |
AT1G61500
|
[S-locus lectin protein kinase family protein] |
-0.454 |
0.095 |
| 113 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.427 |
-0.845 |
| 114 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.412 |
-0.101 |
| 115 |
259558_at |
AT1G21230
|
WAK5, wall associated kinase 5 |
-0.386 |
-1.036 |
| 116 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
-0.382 |
-1.243 |
| 117 |
267077_at |
AT2G40970
|
MYBC1 |
-0.378 |
-0.633 |
| 118 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
-0.375 |
-0.543 |
| 119 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
-0.371 |
0.785 |
| 120 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
-0.365 |
-0.924 |
| 121 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.359 |
0.591 |
| 122 |
259340_at |
AT3G03870
|
unknown |
-0.358 |
0.297 |
| 123 |
264621_at |
AT2G17700
|
STY8, serine/threonine/tyrosine kinase 8 |
-0.358 |
0.442 |
| 124 |
245737_at |
AT1G44160
|
[HSP40/DnaJ peptide-binding protein] |
-0.357 |
-0.446 |
| 125 |
264204_at |
AT1G22710
|
ATSUC2, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2, SUC2, sucrose-proton symporter 2, SUT1, SUCROSE TRANSPORTER 1 |
-0.350 |
0.361 |
| 126 |
258571_at |
AT3G04420
|
anac048, NAC domain containing protein 48, NAC048, NAC domain containing protein 48 |
-0.341 |
-1.268 |
| 127 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.333 |
-0.627 |
| 128 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.329 |
-1.146 |
| 129 |
259520_at |
AT1G12320
|
unknown |
-0.308 |
-0.710 |
| 130 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.308 |
-0.632 |
| 131 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
-0.306 |
-0.095 |
| 132 |
246802_at |
AT5G27000
|
ATK4, kinesin 4, KATD, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA D |
-0.306 |
-0.406 |
| 133 |
266507_at |
AT2G47860
|
SETH6 |
-0.305 |
-1.396 |
| 134 |
264119_at |
AT1G79180
|
ATMYB63, MYB DOMAIN PROTEIN 63, MYB63, myb domain protein 63 |
-0.299 |
0.141 |
| 135 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.296 |
-0.704 |
| 136 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
-0.290 |
0.073 |
| 137 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
-0.289 |
-0.684 |
| 138 |
249007_at |
AT5G44650
|
AtCEST, Arabidopsis thaliana chloroplast protein-enhancing stress tolerance, CEST, chloroplast protein-enhancing stress tolerance, Y3IP1, Ycf3-interacting protein 1 |
-0.286 |
1.064 |
| 139 |
249489_at |
AT5G39090
|
[HXXXD-type acyl-transferase family protein] |
-0.285 |
0.233 |
| 140 |
250031_at |
AT5G18240
|
ATMYR1, ARABIDOPSIS MYB-RELATED PROTEIN 1, MYR1, myb-related protein 1 |
-0.283 |
-1.346 |
| 141 |
261609_at |
AT1G49740
|
[PLC-like phosphodiesterases superfamily protein] |
-0.281 |
0.213 |
| 142 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
-0.281 |
-0.027 |
| 143 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.278 |
-0.227 |
| 144 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.276 |
-1.018 |
| 145 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
-0.273 |
-0.773 |
| 146 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.264 |
-0.429 |
| 147 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
-0.260 |
0.112 |
| 148 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.259 |
-0.203 |
| 149 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.259 |
-0.587 |
| 150 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.258 |
-0.465 |
| 151 |
245906_at |
AT5G11070
|
unknown |
-0.257 |
0.096 |
| 152 |
250028_at |
AT5G18130
|
unknown |
-0.255 |
-0.208 |
| 153 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.253 |
-0.659 |
| 154 |
245186_at |
AT1G67710
|
ARR11, response regulator 11 |
-0.251 |
-0.605 |
| 155 |
254744_at |
AT4G13345
|
MEE55, maternal effect embryo arrest 55 |
-0.249 |
0.133 |
| 156 |
245197_at |
AT1G67800
|
[Copine (Calcium-dependent phospholipid-binding protein) family] |
-0.248 |
0.079 |
| 157 |
247554_at |
AT5G61010
|
ATEXO70E2, exocyst subunit exo70 family protein E2, EXO70E2, exocyst subunit exo70 family protein E2 |
-0.242 |
-0.399 |
| 158 |
245401_at |
AT4G17670
|
unknown |
-0.241 |
0.279 |
| 159 |
258845_at |
AT3G03150
|
unknown |
-0.238 |
0.456 |
| 160 |
256650_at |
AT3G13620
|
PUT4, POLYAMINE UPTAKE TRANSPORTER 4 |
-0.236 |
-0.071 |
| 161 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
-0.236 |
-0.134 |
| 162 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.235 |
-0.343 |
| 163 |
265312_at |
AT2G20240
|
TRM17, TON1 Recruiting Motif 17 |
-0.233 |
0.871 |
| 164 |
247570_at |
AT5G61250
|
AtGUS1, glucuronidase 1, GUS1, glucuronidase 1 |
-0.232 |
0.246 |
| 165 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.232 |
0.198 |
| 166 |
266089_at |
AT2G38010
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.232 |
0.411 |
| 167 |
267182_at |
AT2G23360
|
unknown |
-0.231 |
-0.227 |
| 168 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.224 |
0.414 |
| 169 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
-0.221 |
-0.338 |
| 170 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.220 |
0.577 |
| 171 |
245495_at |
AT4G16400
|
unknown |
-0.216 |
-0.299 |
| 172 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.214 |
0.026 |
| 173 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.214 |
-0.432 |
| 174 |
257031_at |
AT3G19280
|
ATFUT11, FUCT1, FUCTA, FUT11, fucosyltransferase 11 |
-0.213 |
0.179 |
| 175 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.211 |
1.001 |
| 176 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
-0.210 |
0.696 |
| 177 |
262922_at |
AT1G79420
|
unknown |
-0.210 |
-0.382 |
| 178 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.209 |
-0.690 |
| 179 |
247892_at |
AT5G57970
|
[DNA glycosylase superfamily protein] |
-0.208 |
0.630 |
| 180 |
245202_at |
AT1G67720
|
[Leucine-rich repeat protein kinase family protein] |
-0.206 |
-0.481 |
| 181 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
-0.201 |
0.072 |
| 182 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.200 |
-0.091 |
| 183 |
266835_at |
AT2G29990
|
NDA2, alternative NAD(P)H dehydrogenase 2 |
-0.200 |
-0.297 |
| 184 |
262275_at |
AT1G68710
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.199 |
0.528 |
| 185 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.199 |
0.372 |
| 186 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
-0.199 |
0.371 |
| 187 |
264930_at |
AT1G60800
|
AtNIK3, NIK3, NSP-interacting kinase 3 |
-0.196 |
-0.156 |
| 188 |
265320_at |
AT2G18320
|
[F-box associated ubiquitination effector family protein] |
-0.194 |
-0.406 |
| 189 |
260673_at |
AT1G19330
|
unknown |
-0.194 |
0.661 |
| 190 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
-0.194 |
-0.699 |
| 191 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.193 |
-0.830 |
| 192 |
251368_at |
AT3G61380
|
TRM14, TON1 Recruiting Motif 14 |
-0.192 |
-0.144 |
| 193 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
-0.190 |
-0.549 |
| 194 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.190 |
-0.055 |
| 195 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.189 |
0.305 |
| 196 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.188 |
-1.090 |
| 197 |
266812_at |
AT2G44830
|
[Protein kinase superfamily protein] |
-0.188 |
0.391 |
| 198 |
246470_at |
AT5G17080
|
[Cysteine proteinases superfamily protein] |
-0.188 |
-0.319 |
| 199 |
258621_at |
AT3G02830
|
PNT1, PENTA 1, ZFN1, zinc finger protein 1 |
-0.187 |
0.614 |
| 200 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.186 |
-0.225 |
| 201 |
245929_at |
AT5G24760
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.185 |
0.575 |
| 202 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
-0.184 |
0.470 |
| 203 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.184 |
-1.161 |
| 204 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.184 |
-0.918 |
| 205 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
-0.183 |
-0.211 |