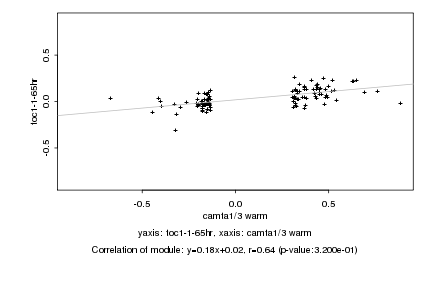
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
camta1/3 warm |
Log2 signal ratio
toc1-1-65hr |
| 1 |
267345_at |
AT2G44240
|
unknown |
0.883 |
-0.014 |
| 2 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.760 |
0.113 |
| 3 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.688 |
0.100 |
| 4 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.648 |
0.228 |
| 5 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.632 |
0.220 |
| 6 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.624 |
0.226 |
| 7 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
0.541 |
0.017 |
| 8 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.529 |
0.119 |
| 9 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.520 |
0.230 |
| 10 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.513 |
0.111 |
| 11 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.499 |
0.166 |
| 12 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.490 |
0.050 |
| 13 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.488 |
0.074 |
| 14 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.479 |
0.135 |
| 15 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.478 |
0.054 |
| 16 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.477 |
-0.027 |
| 17 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.468 |
0.249 |
| 18 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.458 |
0.077 |
| 19 |
245932_at |
AT5G09290
|
[Inositol monophosphatase family protein] |
0.452 |
0.149 |
| 20 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.450 |
0.105 |
| 21 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.446 |
0.078 |
| 22 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.446 |
0.135 |
| 23 |
258203_at |
AT3G13950
|
unknown |
0.437 |
0.194 |
| 24 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.436 |
0.159 |
| 25 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.434 |
0.036 |
| 26 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.430 |
0.181 |
| 27 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.430 |
0.140 |
| 28 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
0.426 |
0.056 |
| 29 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.423 |
0.089 |
| 30 |
259489_at |
AT1G15790
|
unknown |
0.418 |
0.139 |
| 31 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.407 |
0.237 |
| 32 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.379 |
0.139 |
| 33 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.377 |
0.043 |
| 34 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
0.375 |
-0.042 |
| 35 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.370 |
0.135 |
| 36 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.369 |
0.053 |
| 37 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.367 |
-0.070 |
| 38 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.366 |
0.160 |
| 39 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.366 |
0.170 |
| 40 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.359 |
0.052 |
| 41 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.340 |
0.194 |
| 42 |
254571_at |
AT4G19370
|
unknown |
0.340 |
0.110 |
| 43 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.334 |
0.029 |
| 44 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.332 |
0.111 |
| 45 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
0.331 |
0.091 |
| 46 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.330 |
0.024 |
| 47 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.327 |
-0.048 |
| 48 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
0.322 |
0.036 |
| 49 |
263851_at |
AT2G04460
|
unknown |
0.320 |
-0.037 |
| 50 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.319 |
0.124 |
| 51 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.318 |
0.139 |
| 52 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
0.317 |
-0.021 |
| 53 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.316 |
0.266 |
| 54 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.316 |
0.061 |
| 55 |
249987_at |
AT5G18490
|
unknown |
0.313 |
0.039 |
| 56 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
0.312 |
0.044 |
| 57 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.307 |
0.004 |
| 58 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.306 |
-0.059 |
| 59 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.303 |
0.051 |
| 60 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.303 |
0.114 |
| 61 |
250604_at |
AT5G07830
|
AtGUS2, glucuronidase 2, GUS2, glucuronidase 2 |
-0.671 |
0.035 |
| 62 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.448 |
-0.112 |
| 63 |
245910_at |
AT5G09410
|
CAMTA1, calmodulin-binding transcription activator 1, EICBP.B, ethylene induced calmodulin binding protein |
-0.414 |
0.042 |
| 64 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.402 |
0.008 |
| 65 |
246367_at |
AT1G51880
|
RHS6, root hair specific 6 |
-0.399 |
-0.047 |
| 66 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
-0.327 |
-0.028 |
| 67 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.325 |
-0.306 |
| 68 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.320 |
-0.131 |
| 69 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.296 |
-0.054 |
| 70 |
245979_at |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
-0.265 |
-0.009 |
| 71 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
-0.210 |
-0.015 |
| 72 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.206 |
-0.045 |
| 73 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.204 |
0.023 |
| 74 |
259209_at |
AT3G09160
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.203 |
-0.036 |
| 75 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.201 |
-0.038 |
| 76 |
257543_at |
AT3G28960
|
[Transmembrane amino acid transporter family protein] |
-0.200 |
0.092 |
| 77 |
245103_at |
AT2G41590
|
unknown |
-0.199 |
-0.027 |
| 78 |
256646_at |
AT3G13590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.185 |
0.010 |
| 79 |
253659_at |
AT4G30150
|
unknown |
-0.183 |
-0.012 |
| 80 |
251865_at |
AT3G54930
|
[Protein phosphatase 2A regulatory B subunit family protein] |
-0.182 |
-0.048 |
| 81 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
-0.182 |
0.009 |
| 82 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.181 |
-0.106 |
| 83 |
248789_at |
AT5G47440
|
unknown |
-0.180 |
0.005 |
| 84 |
251598_at |
AT3G57600
|
[Integrase-type DNA-binding superfamily protein] |
-0.180 |
-0.071 |
| 85 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.180 |
-0.072 |
| 86 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
-0.179 |
-0.031 |
| 87 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.178 |
-0.012 |
| 88 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.177 |
-0.089 |
| 89 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.171 |
-0.034 |
| 90 |
264179_at |
AT1G02180
|
[ferredoxin-related] |
-0.171 |
0.029 |
| 91 |
252105_at |
AT3G51470
|
[Protein phosphatase 2C family protein] |
-0.169 |
-0.042 |
| 92 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.167 |
-0.035 |
| 93 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.167 |
-0.042 |
| 94 |
252268_at |
AT3G49650
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.166 |
-0.022 |
| 95 |
264971_at |
AT1G67210
|
[Proline-rich spliceosome-associated (PSP) family protein / zinc knuckle (CCHC-type) family protein] |
-0.166 |
0.097 |
| 96 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.165 |
-0.035 |
| 97 |
264378_at |
AT2G25220
|
[Protein kinase superfamily protein] |
-0.161 |
-0.027 |
| 98 |
245322_at |
AT4G14815
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.157 |
0.077 |
| 99 |
253620_at |
AT4G30520
|
SARK, SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE |
-0.156 |
-0.021 |
| 100 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
-0.156 |
-0.111 |
| 101 |
248218_at |
AT5G53710
|
unknown |
-0.154 |
0.086 |
| 102 |
254402_at |
AT4G21310
|
unknown |
-0.153 |
-0.085 |
| 103 |
260493_at |
AT2G41830
|
[Uncharacterized protein] |
-0.153 |
0.021 |
| 104 |
250519_at |
AT5G08460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.152 |
-0.078 |
| 105 |
260960_at |
AT1G05990
|
RHS1, ROOT HAIR SPECIFIC 1 |
-0.151 |
-0.026 |
| 106 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
-0.151 |
0.077 |
| 107 |
251384_at |
AT3G60760
|
unknown |
-0.150 |
0.022 |
| 108 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.147 |
-0.007 |
| 109 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.147 |
0.104 |
| 110 |
258192_at |
AT3G29110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.146 |
0.034 |
| 111 |
263079_at |
AT2G05390
|
[copia-like retrotransposon family, has a 7.2e-200 P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-0.144 |
0.055 |
| 112 |
263962_at |
AT2G36350
|
[Protein kinase superfamily protein] |
-0.144 |
-0.017 |
| 113 |
260441_at |
AT1G68260
|
ALT3, acyl-lipid thioesterase 3 |
-0.142 |
-0.024 |
| 114 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.142 |
-0.035 |
| 115 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.140 |
-0.041 |
| 116 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.137 |
0.026 |
| 117 |
251025_at |
AT5G02190
|
ATASP38, ARABIDOPSIS THALIANA ASPARTIC PROTEASE 38, EMB24, EMBRYO DEFECTIVE 24, PCS1, PROMOTION OF CELL SURVIVAL 1 |
-0.136 |
-0.092 |
| 118 |
259333_at |
AT3G03810
|
EDA30, embryo sac development arrest 30 |
-0.136 |
-0.063 |
| 119 |
261350_at |
AT1G79770
|
unknown |
-0.135 |
0.122 |