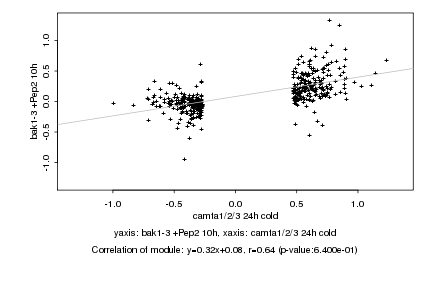
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
camta1/2/3 24h cold |
Log2 signal ratio
bak1-3 +Pep2 10h |
| 1 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
1.229 |
0.684 |
| 2 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
1.137 |
0.474 |
| 3 |
252345_at |
AT3G48640
|
unknown |
1.107 |
0.266 |
| 4 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
1.026 |
0.251 |
| 5 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.969 |
0.315 |
| 6 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.906 |
0.389 |
| 7 |
251419_at |
AT3G60470
|
unknown |
0.899 |
0.035 |
| 8 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.894 |
0.137 |
| 9 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.891 |
0.869 |
| 10 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.891 |
0.698 |
| 11 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.889 |
0.586 |
| 12 |
252346_at |
AT3G48650
|
[pseudogene] |
0.888 |
0.290 |
| 13 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.886 |
0.361 |
| 14 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.883 |
0.229 |
| 15 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.874 |
0.488 |
| 16 |
249004_at |
AT5G44570
|
unknown |
0.856 |
0.152 |
| 17 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.851 |
0.436 |
| 18 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.846 |
0.551 |
| 19 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.844 |
1.251 |
| 20 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.821 |
0.667 |
| 21 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.811 |
0.120 |
| 22 |
249454_at |
AT5G39520
|
unknown |
0.810 |
0.341 |
| 23 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.783 |
0.230 |
| 24 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.776 |
0.652 |
| 25 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.776 |
0.933 |
| 26 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.774 |
0.203 |
| 27 |
249896_at |
AT5G22530
|
unknown |
0.771 |
0.440 |
| 28 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.766 |
0.291 |
| 29 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.763 |
1.344 |
| 30 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.760 |
0.082 |
| 31 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.757 |
0.607 |
| 32 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.754 |
0.520 |
| 33 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.746 |
0.136 |
| 34 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.745 |
0.430 |
| 35 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.744 |
0.221 |
| 36 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.744 |
0.152 |
| 37 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.743 |
0.808 |
| 38 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.740 |
0.603 |
| 39 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.740 |
0.258 |
| 40 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.740 |
0.102 |
| 41 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.730 |
0.083 |
| 42 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.723 |
0.120 |
| 43 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.723 |
0.556 |
| 44 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.718 |
0.485 |
| 45 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.718 |
0.208 |
| 46 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
0.717 |
0.193 |
| 47 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.714 |
0.550 |
| 48 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.713 |
0.722 |
| 49 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.711 |
0.382 |
| 50 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.711 |
0.102 |
| 51 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
0.708 |
0.095 |
| 52 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
0.708 |
0.213 |
| 53 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.706 |
0.246 |
| 54 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.706 |
-0.389 |
| 55 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.704 |
0.528 |
| 56 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.701 |
0.324 |
| 57 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
0.700 |
0.304 |
| 58 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.699 |
0.350 |
| 59 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.696 |
0.226 |
| 60 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.695 |
0.142 |
| 61 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.694 |
0.057 |
| 62 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.694 |
0.312 |
| 63 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.687 |
0.283 |
| 64 |
263182_at |
AT1G05575
|
unknown |
0.686 |
0.349 |
| 65 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.681 |
0.095 |
| 66 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.679 |
0.246 |
| 67 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.676 |
0.401 |
| 68 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
0.664 |
-0.318 |
| 69 |
258203_at |
AT3G13950
|
unknown |
0.663 |
0.515 |
| 70 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.659 |
0.126 |
| 71 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
0.654 |
0.207 |
| 72 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.651 |
0.753 |
| 73 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.651 |
0.113 |
| 74 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.651 |
0.505 |
| 75 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.651 |
0.174 |
| 76 |
259385_at |
AT1G13470
|
unknown |
0.650 |
0.219 |
| 77 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.649 |
0.355 |
| 78 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.648 |
0.863 |
| 79 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.648 |
0.326 |
| 80 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.642 |
0.172 |
| 81 |
257277_at |
AT3G14470
|
[NB-ARC domain-containing disease resistance protein] |
0.642 |
0.279 |
| 82 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.638 |
-0.176 |
| 83 |
255999_at |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
0.637 |
0.196 |
| 84 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.635 |
0.390 |
| 85 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.634 |
0.517 |
| 86 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.633 |
0.550 |
| 87 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.631 |
0.271 |
| 88 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.629 |
0.033 |
| 89 |
263475_at |
AT2G31945
|
unknown |
0.627 |
0.465 |
| 90 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.622 |
0.009 |
| 91 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.620 |
0.198 |
| 92 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.619 |
0.251 |
| 93 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.615 |
0.555 |
| 94 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
0.615 |
0.229 |
| 95 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.615 |
0.244 |
| 96 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.614 |
0.874 |
| 97 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
0.611 |
0.269 |
| 98 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.608 |
0.319 |
| 99 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.607 |
0.275 |
| 100 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.605 |
0.089 |
| 101 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.605 |
0.674 |
| 102 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
0.604 |
0.024 |
| 103 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.603 |
0.667 |
| 104 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.602 |
0.138 |
| 105 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.601 |
0.300 |
| 106 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.601 |
0.578 |
| 107 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
0.600 |
-0.543 |
| 108 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.600 |
0.022 |
| 109 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.599 |
0.617 |
| 110 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.598 |
0.619 |
| 111 |
260117_at |
AT1G33950
|
[Avirulence induced gene (AIG1) family protein] |
0.598 |
0.435 |
| 112 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.598 |
0.040 |
| 113 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.597 |
0.275 |
| 114 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.596 |
0.331 |
| 115 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.595 |
0.311 |
| 116 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.595 |
0.465 |
| 117 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.592 |
0.191 |
| 118 |
261241_at |
AT1G32950
|
[Subtilase family protein] |
0.586 |
0.103 |
| 119 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.585 |
0.418 |
| 120 |
245566_at |
AT4G14610
|
[pseudogene] |
0.579 |
0.244 |
| 121 |
266851_at |
AT2G26820
|
ATPP2-A3, phloem protein 2-A3, PP2-A3, phloem protein 2-A3 |
0.578 |
-0.073 |
| 122 |
245082_at |
AT2G23270
|
unknown |
0.577 |
0.448 |
| 123 |
245076_at |
AT2G23170
|
GH3.3 |
0.577 |
0.228 |
| 124 |
254063_at |
AT4G25390
|
[Protein kinase superfamily protein] |
0.576 |
0.263 |
| 125 |
267345_at |
AT2G44240
|
unknown |
0.572 |
0.046 |
| 126 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.571 |
0.229 |
| 127 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.570 |
0.172 |
| 128 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.570 |
0.464 |
| 129 |
263854_at |
AT2G04430
|
atnudt5, nudix hydrolase homolog 5, NUDT5, nudix hydrolase homolog 5 |
0.567 |
0.254 |
| 130 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.567 |
0.276 |
| 131 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.565 |
0.353 |
| 132 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
0.565 |
0.208 |
| 133 |
261249_at |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
0.564 |
0.341 |
| 134 |
254253_at |
AT4G23320
|
CRK24, cysteine-rich RLK (RECEPTOR-like protein kinase) 24 |
0.563 |
0.180 |
| 135 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.559 |
0.215 |
| 136 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.559 |
0.177 |
| 137 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.558 |
0.035 |
| 138 |
249889_at |
AT5G22540
|
unknown |
0.556 |
0.308 |
| 139 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.555 |
-0.014 |
| 140 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.555 |
0.427 |
| 141 |
254571_at |
AT4G19370
|
unknown |
0.555 |
0.366 |
| 142 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.554 |
0.082 |
| 143 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.554 |
0.651 |
| 144 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.553 |
0.238 |
| 145 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.550 |
0.455 |
| 146 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
0.547 |
0.472 |
| 147 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
0.547 |
0.233 |
| 148 |
260110_at |
AT1G63350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.540 |
0.184 |
| 149 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
0.536 |
0.743 |
| 150 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.536 |
0.381 |
| 151 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.535 |
0.466 |
| 152 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
0.534 |
0.166 |
| 153 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.533 |
0.313 |
| 154 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
0.532 |
0.345 |
| 155 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
0.532 |
0.036 |
| 156 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.531 |
0.133 |
| 157 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
0.530 |
0.092 |
| 158 |
262072_at |
AT1G59590
|
ZCF37 |
0.530 |
0.409 |
| 159 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.527 |
0.051 |
| 160 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.526 |
0.296 |
| 161 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
0.523 |
0.084 |
| 162 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.523 |
0.236 |
| 163 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.523 |
0.156 |
| 164 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.521 |
0.113 |
| 165 |
256470_at |
AT1G42570
|
[pseudogene] |
0.519 |
0.037 |
| 166 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.519 |
0.048 |
| 167 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.518 |
0.215 |
| 168 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
0.518 |
0.175 |
| 169 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
0.516 |
0.170 |
| 170 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.514 |
0.695 |
| 171 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.512 |
0.396 |
| 172 |
258209_at |
AT3G14060
|
unknown |
0.512 |
0.018 |
| 173 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.510 |
0.622 |
| 174 |
257184_at |
AT3G13090
|
ABCC6, ATP-binding cassette C6, ATMRP8, multidrug resistance-associated protein 8, MRP8, multidrug resistance-associated protein 8 |
0.508 |
0.155 |
| 175 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.507 |
0.381 |
| 176 |
255628_at |
AT4G00850
|
GIF3, GRF1-interacting factor 3 |
0.504 |
-0.059 |
| 177 |
259596_at |
AT1G28130
|
GH3.17 |
0.504 |
0.019 |
| 178 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.502 |
0.283 |
| 179 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.501 |
0.207 |
| 180 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.500 |
0.055 |
| 181 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
0.499 |
0.124 |
| 182 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.498 |
0.491 |
| 183 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.497 |
0.136 |
| 184 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.493 |
-0.040 |
| 185 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.493 |
0.017 |
| 186 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.492 |
-0.040 |
| 187 |
254252_at |
AT4G23310
|
CRK23, cysteine-rich RLK (RECEPTOR-like protein kinase) 23 |
0.491 |
0.075 |
| 188 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
0.490 |
0.098 |
| 189 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.490 |
0.195 |
| 190 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.488 |
0.262 |
| 191 |
258351_at |
AT3G17700
|
ATCNGC20, CYCLIC NUCLEOTIDE-GATED CHANNEL 20, CNBT1, cyclic nucleotide-binding transporter 1, CNGC20, cyclic nucleotide gated channel 20 |
0.488 |
0.234 |
| 192 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.487 |
0.007 |
| 193 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.485 |
-0.365 |
| 194 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.484 |
0.552 |
| 195 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.483 |
0.040 |
| 196 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.482 |
0.021 |
| 197 |
264590_at |
AT2G17710
|
unknown |
0.479 |
0.055 |
| 198 |
260734_at |
AT1G17600
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.478 |
0.193 |
| 199 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.478 |
-0.024 |
| 200 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.477 |
0.286 |
| 201 |
254247_at |
AT4G23260
|
CRK18, cysteine-rich RLK (RECEPTOR-like protein kinase) 18 |
0.475 |
0.067 |
| 202 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.474 |
0.160 |
| 203 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.473 |
0.145 |
| 204 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.473 |
0.214 |
| 205 |
245401_at |
AT4G17670
|
unknown |
0.472 |
0.179 |
| 206 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.471 |
0.395 |
| 207 |
261973_at |
AT1G64610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.469 |
0.226 |
| 208 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.468 |
0.055 |
| 209 |
259520_at |
AT1G12320
|
unknown |
0.467 |
0.450 |
| 210 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.467 |
0.196 |
| 211 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.466 |
0.499 |
| 212 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.996 |
-0.018 |
| 213 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.840 |
-0.057 |
| 214 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.724 |
0.054 |
| 215 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.715 |
0.043 |
| 216 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.712 |
0.202 |
| 217 |
250936_at |
AT5G03120
|
unknown |
-0.710 |
-0.307 |
| 218 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
-0.684 |
-0.037 |
| 219 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.682 |
-0.037 |
| 220 |
253346_at |
AT4G33600
|
unknown |
-0.675 |
-0.001 |
| 221 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.671 |
0.069 |
| 222 |
265621_at |
AT2G27300
|
ANAC040, Arabidopsis NAC domain containing protein 40, NTL8, NTM1-like 8 |
-0.666 |
0.103 |
| 223 |
260744_at |
AT1G15010
|
unknown |
-0.661 |
0.344 |
| 224 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.647 |
0.016 |
| 225 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.645 |
-0.081 |
| 226 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.627 |
-0.066 |
| 227 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.622 |
-0.079 |
| 228 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.617 |
-0.080 |
| 229 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.616 |
0.078 |
| 230 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
-0.614 |
0.204 |
| 231 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.592 |
-0.195 |
| 232 |
257672_at |
AT3G20300
|
unknown |
-0.584 |
-0.014 |
| 233 |
245910_at |
AT5G09410
|
CAMTA1, calmodulin-binding transcription activator 1, EICBP.B, ethylene induced calmodulin binding protein |
-0.581 |
0.042 |
| 234 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.570 |
0.139 |
| 235 |
247270_at |
AT5G64220
|
CAMTA2, calmodulin-binding transcription activator 2 |
-0.551 |
0.060 |
| 236 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.548 |
-0.019 |
| 237 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.546 |
0.297 |
| 238 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.545 |
0.012 |
| 239 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
-0.545 |
-0.077 |
| 240 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.531 |
-0.288 |
| 241 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.531 |
-0.047 |
| 242 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.528 |
-0.037 |
| 243 |
250604_at |
AT5G07830
|
AtGUS2, glucuronidase 2, GUS2, glucuronidase 2 |
-0.525 |
0.023 |
| 244 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
-0.519 |
0.307 |
| 245 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.519 |
0.003 |
| 246 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.513 |
-0.107 |
| 247 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.498 |
0.077 |
| 248 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
-0.497 |
0.130 |
| 249 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-0.488 |
0.032 |
| 250 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.487 |
-0.031 |
| 251 |
257057_at |
AT3G15310
|
unknown |
-0.485 |
-0.037 |
| 252 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.482 |
0.269 |
| 253 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.480 |
-0.123 |
| 254 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.479 |
0.068 |
| 255 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.475 |
-0.428 |
| 256 |
260656_at |
AT1G19380
|
unknown |
-0.473 |
-0.058 |
| 257 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.471 |
-0.006 |
| 258 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.466 |
-0.351 |
| 259 |
248968_at |
AT5G45280
|
[Pectinacetylesterase family protein] |
-0.465 |
0.017 |
| 260 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.463 |
-0.032 |
| 261 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.462 |
0.219 |
| 262 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.455 |
-0.019 |
| 263 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
-0.454 |
-0.105 |
| 264 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.454 |
-0.085 |
| 265 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.452 |
-0.280 |
| 266 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.446 |
0.019 |
| 267 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.445 |
-0.104 |
| 268 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.443 |
0.143 |
| 269 |
254098_at |
AT4G25100
|
ATFSD1, ARABIDOPSIS FE SUPEROXIDE DISMUTASE 1, FSD1, Fe superoxide dismutase 1 |
-0.442 |
-0.002 |
| 270 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.442 |
0.031 |
| 271 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.442 |
-0.196 |
| 272 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.440 |
-0.088 |
| 273 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.437 |
0.064 |
| 274 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.437 |
-0.173 |
| 275 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.435 |
0.041 |
| 276 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.435 |
-0.071 |
| 277 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.435 |
-0.159 |
| 278 |
266412_at |
AT2G38720
|
MAP65-5, microtubule-associated protein 65-5 |
-0.434 |
-0.048 |
| 279 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.432 |
0.003 |
| 280 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
-0.430 |
0.089 |
| 281 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.430 |
-0.089 |
| 282 |
245254_at |
AT4G14680
|
APS3 |
-0.430 |
0.068 |
| 283 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.427 |
0.041 |
| 284 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
-0.426 |
-0.104 |
| 285 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.424 |
0.060 |
| 286 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
-0.423 |
-0.009 |
| 287 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.423 |
-0.943 |
| 288 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
-0.422 |
0.010 |
| 289 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.422 |
-0.042 |
| 290 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.421 |
0.122 |
| 291 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.416 |
-0.042 |
| 292 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.410 |
-0.077 |
| 293 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.408 |
0.071 |
| 294 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.407 |
-0.161 |
| 295 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
-0.407 |
-0.189 |
| 296 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.397 |
0.046 |
| 297 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
-0.397 |
-0.183 |
| 298 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-0.394 |
-0.048 |
| 299 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.392 |
-0.408 |
| 300 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.390 |
0.059 |
| 301 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.389 |
-0.342 |
| 302 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.384 |
-0.045 |
| 303 |
247878_at |
AT5G57760
|
unknown |
-0.382 |
-0.593 |
| 304 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.380 |
0.058 |
| 305 |
263457_at |
AT2G22300
|
CAMTA3, CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3, SR1, signal responsive 1 |
-0.380 |
0.115 |
| 306 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
-0.379 |
0.045 |
| 307 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.378 |
-0.053 |
| 308 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.377 |
-0.025 |
| 309 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.375 |
0.026 |
| 310 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.374 |
-0.218 |
| 311 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.374 |
-0.016 |
| 312 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.372 |
-0.346 |
| 313 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.372 |
-0.029 |
| 314 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.371 |
-0.142 |
| 315 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
-0.368 |
-0.001 |
| 316 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.366 |
-0.161 |
| 317 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.366 |
0.029 |
| 318 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.366 |
0.021 |
| 319 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.364 |
-0.062 |
| 320 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
-0.363 |
0.003 |
| 321 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.362 |
-0.284 |
| 322 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.361 |
-0.208 |
| 323 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.359 |
-0.273 |
| 324 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.357 |
0.015 |
| 325 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.352 |
-0.043 |
| 326 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.351 |
-0.085 |
| 327 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.351 |
0.071 |
| 328 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
-0.351 |
0.024 |
| 329 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.350 |
-0.109 |
| 330 |
247455_at |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
-0.350 |
-0.110 |
| 331 |
255899_at |
AT1G17970
|
[RING/U-box superfamily protein] |
-0.349 |
-0.084 |
| 332 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
-0.349 |
0.004 |
| 333 |
245290_at |
AT4G16490
|
[ARM repeat superfamily protein] |
-0.349 |
-0.035 |
| 334 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.348 |
-0.268 |
| 335 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.348 |
-0.148 |
| 336 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.346 |
-0.195 |
| 337 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.345 |
-0.004 |
| 338 |
260982_at |
AT1G53520
|
AtFAP3, FAP3, fatty-acid-binding protein 3 |
-0.345 |
-0.383 |
| 339 |
263689_at |
AT1G26820
|
RNS3, ribonuclease 3 |
-0.345 |
-0.014 |
| 340 |
253679_at |
AT4G29610
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.344 |
-0.054 |
| 341 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
-0.344 |
-0.150 |
| 342 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.344 |
0.020 |
| 343 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.343 |
-0.247 |
| 344 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.343 |
0.101 |
| 345 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.341 |
-0.027 |
| 346 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
-0.340 |
-0.085 |
| 347 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
-0.340 |
-0.040 |
| 348 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.336 |
-0.157 |
| 349 |
250560_at |
AT5G08020
|
ATRPA70B, ARABIDOPSIS THALIANA RPA70-KDA SUBUNIT B, RPA70B, RPA70-kDa subunit B |
-0.336 |
-0.077 |
| 350 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.335 |
-0.021 |
| 351 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
-0.333 |
-0.036 |
| 352 |
248139_at |
AT5G54970
|
unknown |
-0.332 |
-0.080 |
| 353 |
267112_at |
AT2G14750
|
AKN1, APS KINASE 1, APK, APS kinase, APK1, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 1, ATAKN1 |
-0.331 |
0.023 |
| 354 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.329 |
-0.071 |
| 355 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
-0.327 |
0.004 |
| 356 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.326 |
-0.052 |
| 357 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
-0.324 |
0.017 |
| 358 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-0.322 |
0.247 |
| 359 |
256646_at |
AT3G13590
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.322 |
0.081 |
| 360 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
-0.319 |
-0.124 |
| 361 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
-0.319 |
-0.249 |
| 362 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.319 |
0.046 |
| 363 |
251024_at |
AT5G02180
|
[Transmembrane amino acid transporter family protein] |
-0.318 |
-0.011 |
| 364 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
-0.318 |
0.018 |
| 365 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.316 |
-0.194 |
| 366 |
260441_at |
AT1G68260
|
ALT3, acyl-lipid thioesterase 3 |
-0.316 |
-0.049 |
| 367 |
260389_at |
AT1G74055
|
unknown |
-0.314 |
-0.070 |
| 368 |
245513_at |
AT4G15780
|
ATVAMP724, vesicle-associated membrane protein 724, VAMP724, vesicle-associated membrane protein 724 |
-0.314 |
-0.016 |
| 369 |
263535_at |
AT2G24970
|
unknown |
-0.314 |
-0.120 |
| 370 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.313 |
-0.046 |
| 371 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.313 |
0.126 |
| 372 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
-0.311 |
-0.052 |
| 373 |
258630_at |
AT3G02820
|
[zinc knuckle (CCHC-type) family protein] |
-0.311 |
-0.044 |
| 374 |
260385_at |
AT1G74090
|
ATSOT18, DESULFO-GLUCOSINOLATE SULFOTRANSFERASE 18, ATST5B, ARABIDOPSIS SULFOTRANSFERASE 5B, SOT18, desulfo-glucosinolate sulfotransferase 18 |
-0.310 |
-0.101 |
| 375 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.306 |
-0.090 |
| 376 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-0.306 |
-0.231 |
| 377 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.305 |
0.020 |
| 378 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.305 |
-0.012 |
| 379 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.305 |
-0.144 |
| 380 |
246860_at |
AT5G25840
|
unknown |
-0.305 |
0.062 |
| 381 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.302 |
-0.079 |
| 382 |
247399_at |
AT5G62960
|
unknown |
-0.302 |
0.038 |
| 383 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.301 |
-0.076 |
| 384 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-0.301 |
-0.036 |
| 385 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.299 |
-0.233 |
| 386 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.299 |
-0.184 |
| 387 |
261422_at |
AT1G18730
|
NDF6, NDH dependent flow 6, PnsB4, Photosynthetic NDH subcomplex B 4 |
-0.297 |
-0.085 |
| 388 |
265695_at |
AT2G24490
|
ATRPA2, ATRPA32A, ROR1, SUPPRESSOR OF ROS1, RPA2, replicon protein A2, RPA32A |
-0.297 |
-0.078 |
| 389 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.297 |
-0.121 |
| 390 |
263130_at |
AT1G78650
|
POLD3 |
-0.295 |
-0.042 |
| 391 |
245635_at |
AT1G25250
|
AtIDD16, indeterminate(ID)-domain 16, IDD16, indeterminate(ID)-domain 16 |
-0.294 |
-0.013 |
| 392 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
-0.293 |
-0.030 |
| 393 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
-0.292 |
-0.079 |
| 394 |
256418_at |
AT3G06160
|
[AP2/B3-like transcriptional factor family protein] |
-0.291 |
0.016 |
| 395 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.291 |
0.614 |
| 396 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
-0.290 |
-0.109 |
| 397 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
-0.290 |
-0.156 |
| 398 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
-0.290 |
-0.268 |
| 399 |
259207_at |
AT3G09050
|
unknown |
-0.289 |
-0.049 |
| 400 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.287 |
-0.224 |
| 401 |
256284_at |
AT3G12530
|
PSF2 |
-0.285 |
-0.108 |
| 402 |
263477_at |
AT2G31790
|
[UDP-Glycosyltransferase superfamily protein] |
-0.284 |
-0.083 |
| 403 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.283 |
-0.107 |
| 404 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.283 |
0.336 |
| 405 |
267455_at |
AT2G33760
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.282 |
-0.036 |
| 406 |
251961_at |
AT3G53620
|
AtPPa4, pyrophosphorylase 4, PPa4, pyrophosphorylase 4 |
-0.282 |
-0.092 |
| 407 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.282 |
0.033 |
| 408 |
249340_at |
AT5G41140
|
[Myosin heavy chain-related protein] |
-0.282 |
-0.178 |
| 409 |
245864_at |
AT1G58070
|
unknown |
-0.281 |
-0.045 |
| 410 |
266946_at |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.281 |
-0.004 |
| 411 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.281 |
0.321 |
| 412 |
248074_at |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
-0.281 |
-0.456 |
| 413 |
260877_at |
AT1G21500
|
unknown |
-0.279 |
-0.215 |
| 414 |
259232_at |
AT3G11420
|
unknown |
-0.279 |
0.043 |
| 415 |
253883_at |
AT4G27660
|
unknown |
-0.278 |
-0.132 |
| 416 |
254697_at |
AT4G17970
|
ALMT12, aluminum-activated, malate transporter 12, ATALMT12, QUAC1, quick-activating anion channel 1 |
-0.278 |
0.106 |
| 417 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.278 |
0.092 |
| 418 |
246315_at |
AT3G56870
|
unknown |
-0.277 |
-0.040 |
| 419 |
254703_at |
AT4G17960
|
unknown |
-0.277 |
-0.059 |
| 420 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.276 |
-0.080 |
| 421 |
261175_at |
AT1G04800
|
[glycine-rich protein] |
-0.276 |
-0.042 |