|
probeID |
AGICode |
Annotation |
Log2 signal ratio
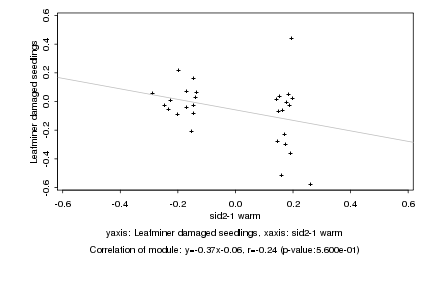
sid2-1 warm |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
0.260 |
-0.572 |
| 2 |
252857_at |
AT4G39756
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.197 |
0.028 |
| 3 |
264774_at |
AT1G22890
|
unknown |
0.192 |
0.444 |
| 4 |
267158_at |
AT2G37640
|
ATEXP3, EXPANSIN 3, ATEXPA3, ARABIDOPSIS THALIANA EXPANSIN A3, ATHEXP ALPHA 1.9, EXP3 |
0.190 |
-0.358 |
| 5 |
252962_at |
AT4G38780
|
[Pre-mRNA-processing-splicing factor] |
0.187 |
-0.022 |
| 6 |
253931_at |
AT4G26770
|
[Phosphatidate cytidylyltransferase family protein] |
0.181 |
0.051 |
| 7 |
260617_at |
AT1G53345
|
unknown |
0.177 |
-0.000 |
| 8 |
266295_at |
AT2G29550
|
TUB7, tubulin beta-7 chain |
0.171 |
-0.297 |
| 9 |
266117_at |
AT2G02170
|
[Remorin family protein] |
0.169 |
-0.225 |
| 10 |
263973_at |
AT2G42740
|
RPL16A, ribosomal protein large subunit 16A |
0.161 |
-0.058 |
| 11 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.158 |
-0.513 |
| 12 |
261406_at |
AT1G18800
|
NRP2, NAP1-related protein 2 |
0.150 |
0.040 |
| 13 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
0.149 |
-0.064 |
| 14 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
0.145 |
-0.276 |
| 15 |
263286_at |
AT2G36160
|
[Ribosomal protein S11 family protein] |
0.141 |
0.021 |
| 16 |
256391_at |
AT3G06090
|
unknown |
-0.290 |
0.062 |
| 17 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
-0.249 |
-0.024 |
| 18 |
257491_at |
AT1G06170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.233 |
-0.052 |
| 19 |
264502_at |
AT1G09400
|
[FMN-linked oxidoreductases superfamily protein] |
-0.229 |
0.008 |
| 20 |
262810_at |
AT1G11710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.203 |
-0.088 |
| 21 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.199 |
0.220 |
| 22 |
258202_at |
AT3G13940
|
[DNA binding] |
-0.172 |
0.072 |
| 23 |
253370_at |
AT4G33230
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.172 |
-0.037 |
| 24 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
-0.156 |
-0.208 |
| 25 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-0.148 |
0.162 |
| 26 |
248006_at |
AT5G56250
|
HAP8, HAPLESS 8 |
-0.148 |
-0.080 |
| 27 |
248240_at |
AT5G53950
|
ANAC098, Arabidopsis NAC domain containing protein 98, ATCUC2, CUP-SHAPED COTYLEDON 2, CUC2, CUP-SHAPED COTYLEDON 2 |
-0.146 |
-0.027 |
| 28 |
248593_at |
AT5G49180
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.141 |
0.034 |
| 29 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
-0.137 |
0.069 |