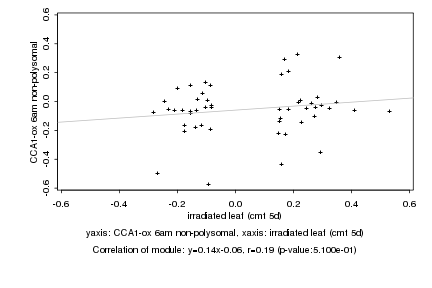
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
irradiated leaf (cmt 5d) |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.530 |
-0.063 |
| 2 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.408 |
-0.059 |
| 3 |
265327_at |
AT2G18210
|
unknown |
0.358 |
0.311 |
| 4 |
257478_at |
AT1G16130
|
WAKL2, wall associated kinase-like 2 |
0.346 |
-0.003 |
| 5 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
0.324 |
-0.043 |
| 6 |
257076_at |
AT3G19680
|
unknown |
0.295 |
-0.024 |
| 7 |
252681_at |
AT3G44350
|
anac061, NAC domain containing protein 61, NAC061, NAC domain containing protein 61 |
0.291 |
-0.353 |
| 8 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.281 |
0.028 |
| 9 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.275 |
-0.035 |
| 10 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.272 |
-0.101 |
| 11 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.262 |
-0.009 |
| 12 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.245 |
-0.043 |
| 13 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.227 |
-0.140 |
| 14 |
246926_at |
AT5G25240
|
unknown |
0.224 |
0.008 |
| 15 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.215 |
-0.004 |
| 16 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.213 |
0.328 |
| 17 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.183 |
-0.055 |
| 18 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
0.183 |
0.209 |
| 19 |
245272_at |
AT4G17250
|
unknown |
0.171 |
-0.228 |
| 20 |
248199_at |
AT5G54170
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.167 |
0.294 |
| 21 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.158 |
0.192 |
| 22 |
252652_at |
AT3G44720
|
ADT4, arogenate dehydratase 4 |
0.156 |
-0.436 |
| 23 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.155 |
-0.112 |
| 24 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.149 |
-0.049 |
| 25 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.149 |
-0.132 |
| 26 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
0.148 |
-0.219 |
| 27 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.286 |
-0.074 |
| 28 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.270 |
-0.493 |
| 29 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.245 |
0.004 |
| 30 |
261247_at |
AT1G20070
|
unknown |
-0.232 |
-0.052 |
| 31 |
258807_at |
AT3G04030
|
MYR2 |
-0.212 |
-0.060 |
| 32 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.200 |
0.092 |
| 33 |
256066_at |
AT1G06980
|
unknown |
-0.183 |
-0.056 |
| 34 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.179 |
-0.164 |
| 35 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.177 |
-0.204 |
| 36 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.157 |
0.112 |
| 37 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.157 |
-0.065 |
| 38 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.156 |
-0.079 |
| 39 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.140 |
-0.178 |
| 40 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
-0.137 |
-0.062 |
| 41 |
260639_at |
AT1G53180
|
unknown |
-0.132 |
0.019 |
| 42 |
256923_at |
AT3G29635
|
[HXXXD-type acyl-transferase family protein] |
-0.118 |
-0.162 |
| 43 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
-0.114 |
0.058 |
| 44 |
249296_at |
AT5G41310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.105 |
-0.036 |
| 45 |
245330_at |
AT4G14930
|
[Survival protein SurE-like phosphatase/nucleotidase] |
-0.104 |
0.138 |
| 46 |
257444_at |
AT2G12550
|
NUB1, homolog of human NUB1 |
-0.099 |
0.009 |
| 47 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.095 |
-0.568 |
| 48 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.087 |
-0.188 |
| 49 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.087 |
0.112 |
| 50 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.086 |
-0.021 |
| 51 |
251224_at |
AT3G62620
|
[sucrose-phosphatase-related] |
-0.086 |
-0.035 |