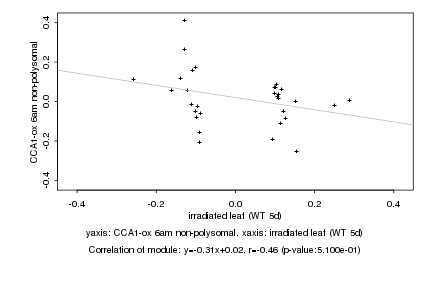
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
irradiated leaf (WT 5d) |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.286 |
0.008 |
| 2 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.250 |
-0.018 |
| 3 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.152 |
-0.251 |
| 4 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.151 |
0.005 |
| 5 |
260204_at |
AT1G52900
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.126 |
-0.082 |
| 6 |
254614_at |
AT4G19191
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.119 |
-0.046 |
| 7 |
247214_at |
AT5G64850
|
unknown |
0.116 |
0.065 |
| 8 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.112 |
-0.107 |
| 9 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
0.108 |
0.038 |
| 10 |
256707_at |
AT3G30310
|
unknown |
0.108 |
0.016 |
| 11 |
253055_at |
AT4G37630
|
CYCD5;1, cyclin d5;1 |
0.105 |
0.027 |
| 12 |
245246_at |
AT1G44224
|
[ECA1 gametogenesis related family protein] |
0.103 |
0.091 |
| 13 |
259425_at |
AT1G01460
|
ATPIPK11, PIPK11 |
0.100 |
0.072 |
| 14 |
254976_at |
AT4G10510
|
[Subtilase family protein] |
0.098 |
0.041 |
| 15 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
0.097 |
0.075 |
| 16 |
261460_at |
AT1G07880
|
ATMPK13 |
0.092 |
-0.191 |
| 17 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.259 |
0.115 |
| 18 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.163 |
0.059 |
| 19 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.139 |
0.120 |
| 20 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
-0.130 |
0.416 |
| 21 |
251232_at |
AT3G62780
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.129 |
0.268 |
| 22 |
260472_at |
AT1G10990
|
unknown |
-0.123 |
0.059 |
| 23 |
264939_at |
AT1G60630
|
[Leucine-rich repeat protein kinase family protein] |
-0.113 |
-0.014 |
| 24 |
252054_at |
AT3G52540
|
ATOFP18, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 18, OFP18, ovate family protein 18 |
-0.109 |
0.160 |
| 25 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
-0.103 |
0.177 |
| 26 |
246614_at |
AT5G35410
|
ATSOS2, SALT OVERLY SENSITIVE 2, CIPK24, CBL-INTERACTING PROTEIN KINASE 24, SNRK3.11, SNF1-RELATED PROTEIN KINASE 3.11, SOS2, SALT OVERLY SENSITIVE 2 |
-0.103 |
-0.046 |
| 27 |
252993_at |
AT4G38540
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.100 |
-0.077 |
| 28 |
249092_at |
AT5G43710
|
[Glycosyl hydrolase family 47 protein] |
-0.098 |
-0.025 |
| 29 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.093 |
-0.204 |
| 30 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.092 |
-0.155 |
| 31 |
249279_at |
AT5G41920
|
[GRAS family transcription factor] |
-0.089 |
-0.059 |