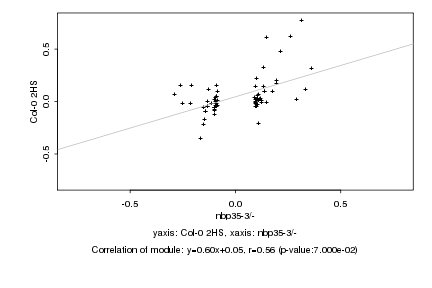
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
nbp35-3/- |
Log2 signal ratio
Col-0 2HS |
| 1 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
0.358 |
0.322 |
| 2 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.328 |
0.121 |
| 3 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.310 |
0.782 |
| 4 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.287 |
0.025 |
| 5 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
0.261 |
0.627 |
| 6 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.210 |
0.481 |
| 7 |
263851_at |
AT2G04460
|
unknown |
0.193 |
0.204 |
| 8 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
0.191 |
0.175 |
| 9 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.172 |
0.098 |
| 10 |
247341_at |
AT5G63720
|
KPL, KOKOPELLI |
0.146 |
-0.005 |
| 11 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.145 |
0.618 |
| 12 |
257664_at |
AT3G20400
|
EMB2743, EMBRYO DEFECTIVE 2743 |
0.136 |
0.103 |
| 13 |
257263_at |
AT3G22070
|
[proline-rich family protein] |
0.134 |
0.097 |
| 14 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
0.129 |
0.144 |
| 15 |
256014_at |
AT1G19200
|
unknown |
0.129 |
0.330 |
| 16 |
246386_at |
AT1G77410
|
BGAL16, beta-galactosidase 16 |
0.123 |
-0.009 |
| 17 |
245741_at |
AT1G44120
|
CSI2, CELLULOSE SYNTHASE INTERACTIVE 2 |
0.121 |
0.010 |
| 18 |
245650_at |
AT1G24735
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.116 |
0.034 |
| 19 |
256693_at |
AT3G32060
|
[Mutator-like transposase family, has a 4.0e-63 P-value blast match to O80466 /172-336 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.110 |
0.023 |
| 20 |
256194_at |
AT1G36810
|
[pseudogene] |
0.110 |
0.020 |
| 21 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.109 |
-0.204 |
| 22 |
253769_at |
AT4G28560
|
RIC7, ROP-interactive CRIB motif-containing protein 7 |
0.107 |
0.019 |
| 23 |
262127_at |
AT1G52550
|
unknown |
0.105 |
0.069 |
| 24 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
0.104 |
0.063 |
| 25 |
255545_at |
AT4G01890
|
[Pectin lyase-like superfamily protein] |
0.103 |
0.037 |
| 26 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.102 |
-0.022 |
| 27 |
248205_at |
AT5G54300
|
unknown |
0.098 |
-0.041 |
| 28 |
255993_at |
AT1G29770
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.097 |
-0.019 |
| 29 |
255174_at |
AT4G08030
|
[gypsy-like retrotransposon family (Athila), has a 1.5e-66 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
0.097 |
0.025 |
| 30 |
253044_at |
AT4G37290
|
unknown |
0.096 |
0.221 |
| 31 |
264311_at |
AT1G70400
|
unknown |
0.096 |
0.021 |
| 32 |
258058_at |
AT3G28980
|
unknown |
0.095 |
-0.018 |
| 33 |
248408_at |
AT5G51520
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.094 |
0.008 |
| 34 |
261243_at |
AT1G20180
|
unknown |
0.094 |
0.023 |
| 35 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
0.094 |
0.146 |
| 36 |
256031_at |
AT1G34100
|
[choline kinase, putative, similar to choline kinase (GmCK2p) GI:1438881 from (Glycine max)] |
0.093 |
-0.046 |
| 37 |
264559_at |
AT1G09610
|
GXM3, glucuronoxylan methyltransferase 3 |
0.091 |
-0.009 |
| 38 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
0.089 |
0.046 |
| 39 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
-0.291 |
0.067 |
| 40 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.265 |
0.158 |
| 41 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
-0.255 |
-0.017 |
| 42 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
-0.217 |
-0.018 |
| 43 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.211 |
0.161 |
| 44 |
266222_at |
AT2G28780
|
unknown |
-0.170 |
-0.346 |
| 45 |
259661_at |
AT1G55265
|
unknown |
-0.156 |
-0.050 |
| 46 |
261247_at |
AT1G20070
|
unknown |
-0.156 |
-0.217 |
| 47 |
259394_at |
AT1G06420
|
unknown |
-0.149 |
-0.162 |
| 48 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.143 |
-0.086 |
| 49 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
-0.143 |
-0.087 |
| 50 |
251527_at |
AT3G58650
|
TRM7, TON1 Recruiting Motif 7 |
-0.137 |
-0.042 |
| 51 |
265767_at |
AT2G48110
|
MED33B, REF4, REDUCED EPIDERMAL FLUORESCENCE 4 |
-0.135 |
-0.045 |
| 52 |
260032_at |
AT1G68750
|
ATPPC4, phosphoenolpyruvate carboxylase 4, PPC4, phosphoenolpyruvate carboxylase 4 |
-0.133 |
0.004 |
| 53 |
250998_at |
AT5G02620
|
ANK1, ankyrin-like1, ATANK1 |
-0.128 |
0.115 |
| 54 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
-0.117 |
-0.014 |
| 55 |
247883_at |
AT5G57790
|
unknown |
-0.104 |
0.024 |
| 56 |
249212_at |
AT5G42690
|
unknown |
-0.104 |
-0.070 |
| 57 |
254186_at |
AT4G24010
|
ATCSLG1, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G1, CSLG1, cellulose synthase like G1 |
-0.104 |
-0.049 |
| 58 |
247554_at |
AT5G61010
|
ATEXO70E2, exocyst subunit exo70 family protein E2, EXO70E2, exocyst subunit exo70 family protein E2 |
-0.102 |
-0.082 |
| 59 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.102 |
-0.051 |
| 60 |
264062_at |
AT2G27950
|
[Ring/U-Box superfamily protein] |
-0.101 |
-0.055 |
| 61 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
-0.100 |
-0.119 |
| 62 |
254618_at |
AT4G18780
|
ATCESA8, CELLULOSE SYNTHASE 8, CESA8, CELLULOSE SYNTHASE 8, IRX1, IRREGULAR XYLEM 1, LEW2, LEAF WILTING 2 |
-0.098 |
0.001 |
| 63 |
263003_at |
AT1G54450
|
[Calcium-binding EF-hand family protein] |
-0.097 |
-0.035 |
| 64 |
258865_at |
AT3G03200
|
anac045, NAC domain containing protein 45, NAC045, NAC domain containing protein 45 |
-0.097 |
0.045 |
| 65 |
264281_at |
AT1G61830
|
[pseudogene] |
-0.097 |
-0.012 |
| 66 |
262081_at |
AT1G59540
|
ZCF125 |
-0.095 |
-0.015 |
| 67 |
247003_at |
AT5G67550
|
unknown |
-0.094 |
-0.039 |
| 68 |
261741_at |
AT1G47870
|
ATE2F2, ATE2FC, ARABIDOPSIS THALIANA HOMOLOG OF E2F C, E2FC |
-0.093 |
-0.029 |
| 69 |
257359_x_at |
AT2G34290
|
[Protein kinase superfamily protein] |
-0.093 |
-0.013 |
| 70 |
246235_at |
AT4G36830
|
HOS3-1 |
-0.092 |
0.054 |
| 71 |
245068_at |
AT2G23260
|
UGT84B1, UDP-glucosyl transferase 84B1 |
-0.092 |
0.051 |
| 72 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
-0.091 |
0.155 |
| 73 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
-0.089 |
0.019 |
| 74 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.089 |
-0.033 |
| 75 |
247742_at |
AT5G58980
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.087 |
0.098 |