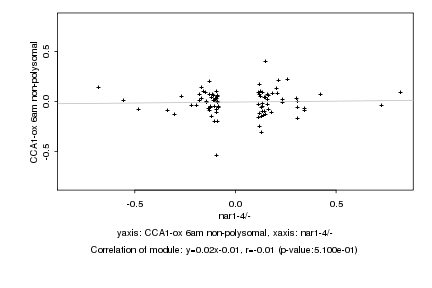
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
nar1-4/- |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.818 |
0.091 |
| 2 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.721 |
-0.032 |
| 3 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.419 |
0.072 |
| 4 |
263515_at |
AT2G21640
|
unknown |
0.342 |
-0.067 |
| 5 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.340 |
-0.081 |
| 6 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
0.308 |
-0.056 |
| 7 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.308 |
-0.167 |
| 8 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.305 |
0.003 |
| 9 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.302 |
0.035 |
| 10 |
260522_x_at |
AT2G41730
|
unknown |
0.257 |
0.225 |
| 11 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.233 |
0.028 |
| 12 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.230 |
-0.003 |
| 13 |
264868_at |
AT1G24095
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.209 |
0.217 |
| 14 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.204 |
0.085 |
| 15 |
247515_at |
AT5G61740
|
ABCA10, ATP-binding cassette A10, ATATH14, ARABIDOPSIS THALIANA ABC2 HOMOLOG 14, ATH14, ABC2 homolog 14 |
0.203 |
0.133 |
| 16 |
245029_at |
AT2G26580
|
YAB5, YABBY5 |
0.181 |
0.081 |
| 17 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.179 |
-0.106 |
| 18 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.160 |
-0.072 |
| 19 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.160 |
0.065 |
| 20 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
0.157 |
-0.023 |
| 21 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
0.156 |
0.076 |
| 22 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.155 |
0.021 |
| 23 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.148 |
0.409 |
| 24 |
251001_at |
AT5G02670
|
unknown |
0.146 |
0.044 |
| 25 |
263214_at |
AT1G30660
|
[nucleic acid binding] |
0.146 |
-0.120 |
| 26 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.141 |
-0.094 |
| 27 |
248432_at |
AT5G51390
|
unknown |
0.141 |
0.041 |
| 28 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
0.138 |
-0.133 |
| 29 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
0.134 |
-0.019 |
| 30 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
0.134 |
-0.095 |
| 31 |
250660_at |
AT5G07060
|
MAC5C, MOS4-associated complex subunit 5C |
0.131 |
0.090 |
| 32 |
262127_at |
AT1G52550
|
unknown |
0.130 |
-0.041 |
| 33 |
249402_at |
AT5G40155
|
[Defensin-like (DEFL) family protein] |
0.126 |
-0.144 |
| 34 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.125 |
-0.051 |
| 35 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.125 |
-0.304 |
| 36 |
253671_at |
AT4G29990
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.124 |
0.106 |
| 37 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.124 |
0.058 |
| 38 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.118 |
0.179 |
| 39 |
247093_at |
AT5G66350
|
SHI, SHORT INTERNODES |
0.118 |
-0.116 |
| 40 |
247737_at |
AT5G59200
|
OTP80, ORGANELLE TRANSCRIPT PROCESSING 80 |
0.117 |
-0.242 |
| 41 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.117 |
0.071 |
| 42 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.113 |
-0.027 |
| 43 |
251051_at |
AT5G02460
|
[Dof-type zinc finger DNA-binding family protein] |
0.112 |
0.095 |
| 44 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.111 |
-0.156 |
| 45 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
-0.684 |
0.146 |
| 46 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.558 |
0.018 |
| 47 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
-0.483 |
-0.079 |
| 48 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.338 |
-0.088 |
| 49 |
245496_at |
AT4G16440
|
GOLLUM, Growth at different Oxygen Levels Influences Morphogenesis, NAR1, homolog of yeast NAR1 |
-0.303 |
-0.122 |
| 50 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.272 |
0.058 |
| 51 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.220 |
-0.032 |
| 52 |
260093_at |
AT1G73270
|
scpl6, serine carboxypeptidase-like 6 |
-0.197 |
-0.034 |
| 53 |
245037_at |
AT2G26420
|
PIP5K3, 1-phosphatidylinositol-4-phosphate 5-kinase 3 |
-0.181 |
0.079 |
| 54 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
-0.179 |
0.018 |
| 55 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.172 |
0.036 |
| 56 |
249438_at |
AT5G40010
|
AATP1, AAA-ATPase 1, ASD, ATPase-in-Seed-Development |
-0.169 |
0.145 |
| 57 |
266549_at |
AT2G35150
|
EXL7, EXORDIUM LIKE 7 |
-0.162 |
0.101 |
| 58 |
267276_at |
AT2G30130
|
ASL5, LBD12, PCK1, PEACOCK 1 |
-0.151 |
0.095 |
| 59 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.146 |
-0.001 |
| 60 |
259028_at |
AT3G09290
|
TAC1, telomerase activator1 |
-0.144 |
0.001 |
| 61 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.134 |
-0.062 |
| 62 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.132 |
-0.089 |
| 63 |
258578_at |
AT3G04200
|
[RmlC-like cupins superfamily protein] |
-0.131 |
0.080 |
| 64 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
-0.130 |
0.200 |
| 65 |
262305_at |
AT1G70950
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.126 |
-0.045 |
| 66 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.125 |
-0.056 |
| 67 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.119 |
-0.148 |
| 68 |
263775_at |
AT2G46410
|
CPC, CAPRICE |
-0.119 |
0.044 |
| 69 |
265439_at |
AT2G21045
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.116 |
0.076 |
| 70 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.112 |
0.016 |
| 71 |
245725_at |
AT1G73370
|
ATSUS6, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 6, SUS6, sucrose synthase 6 |
-0.109 |
0.069 |
| 72 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.108 |
0.014 |
| 73 |
260702_at |
AT1G32250
|
[Calcium-binding EF-hand family protein] |
-0.108 |
-0.044 |
| 74 |
247318_at |
AT5G63990
|
[Inositol monophosphatase family protein] |
-0.105 |
-0.196 |
| 75 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
-0.104 |
0.039 |
| 76 |
259142_at |
AT3G10200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.099 |
0.105 |
| 77 |
263373_at |
AT2G20515
|
unknown |
-0.098 |
0.034 |
| 78 |
257512_at |
AT1G35250
|
ALT2, acyl-lipid thioesterase 2 |
-0.096 |
-0.102 |
| 79 |
251550_at |
AT3G58800
|
unknown |
-0.096 |
-0.532 |
| 80 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.095 |
-0.079 |
| 81 |
265685_at |
AT2G24430
|
ANAC038, NAC domain containing protein 38, ANAC039, Arabidopsis NAC domain containing protein 39, NAC038, NAC domain containing protein 38 |
-0.094 |
0.051 |
| 82 |
264242_at |
AT1G54640
|
[F-box family protein-related] |
-0.094 |
0.069 |
| 83 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
-0.094 |
-0.008 |
| 84 |
248584_at |
AT5G49960
|
unknown |
-0.090 |
0.001 |
| 85 |
266177_at |
AT2G02270
|
ATPP2-B3 |
-0.090 |
-0.194 |
| 86 |
260911_at |
AT1G02490
|
unknown |
-0.089 |
-0.055 |
| 87 |
256829_at |
AT3G22850
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.089 |
-0.046 |