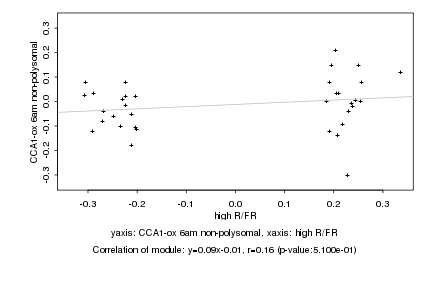
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
high R/FR |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
255159_at |
AT4G07810
|
[copia-like retrotransposon family, has a 7.1e-230 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.335 |
0.122 |
| 2 |
259999_at |
AT1G68080
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.256 |
0.079 |
| 3 |
254714_at |
AT4G13480
|
AtMYB79, myb domain protein 79, MYB79, myb domain protein 79 |
0.254 |
0.001 |
| 4 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.250 |
0.151 |
| 5 |
245944_at |
AT5G19520
|
ATMSL9, MSL9, mechanosensitive channel of small conductance-like 9 |
0.243 |
0.007 |
| 6 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.238 |
-0.019 |
| 7 |
254364_at |
AT4G22020
|
[pseudogene] |
0.236 |
-0.006 |
| 8 |
262352_at |
AT1G64250
|
[Mutator-like transposase family, has a 2.2e-20 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.230 |
-0.038 |
| 9 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.226 |
-0.300 |
| 10 |
258263_at |
AT3G15780
|
unknown |
0.216 |
-0.091 |
| 11 |
252627_at |
AT3G44950
|
[glycine-rich protein] |
0.208 |
0.033 |
| 12 |
246848_at |
AT5G26840
|
unknown |
0.207 |
-0.137 |
| 13 |
263290_at |
AT2G10930
|
unknown |
0.204 |
0.033 |
| 14 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.203 |
0.211 |
| 15 |
252755_at |
AT3G43530
|
unknown |
0.194 |
0.148 |
| 16 |
263059_at |
AT2G07660
|
[gypsy-like retrotransposon family, has a 2.8e-303 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.190 |
0.078 |
| 17 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
0.190 |
-0.119 |
| 18 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
0.185 |
0.004 |
| 19 |
256147_at |
AT1G55080
|
MED9 |
-0.309 |
0.025 |
| 20 |
264115_at |
AT2G31290
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.306 |
0.080 |
| 21 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.292 |
-0.121 |
| 22 |
264610_at |
AT1G04645
|
[Plant self-incompatibility protein S1 family] |
-0.289 |
0.034 |
| 23 |
257047_at |
AT3G19570
|
QWRF1, QWRF domain containing 1, SCO3, SNOWY COTYLEDON 3 |
-0.271 |
-0.081 |
| 24 |
247749_at |
AT5G58850
|
ATMYB119, MYB DOMAIN PROTEIN 119, MYB119, myb domain protein 119 |
-0.269 |
-0.039 |
| 25 |
259631_at |
AT1G56410
|
ERD2, EARLY-RESPONSIVE TO DEHYDRATION 2, HSP70T-1, HEAT SHOCK PROTEIN 70T-1 |
-0.250 |
-0.059 |
| 26 |
264401_at |
AT1G61720
|
BAN, BANYULS |
-0.234 |
-0.102 |
| 27 |
250492_at |
AT5G09790
|
ATXR5, ARABIDOPSIS TRITHORAX-RELATED PROTEIN 5, PDE336, PIGMENT DEFECTIVE 336, SDG15, SETDOMAIN GROUP 15 |
-0.230 |
0.012 |
| 28 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.225 |
0.081 |
| 29 |
245467_at |
AT4G16610
|
[C2H2-like zinc finger protein] |
-0.224 |
-0.015 |
| 30 |
256282_at |
AT3G12550
|
FDM3, factor of DNA methylation 3 |
-0.224 |
0.023 |
| 31 |
255352_at |
AT4G03860
|
[transposon protein -related, similar to athila retroelement] |
-0.213 |
-0.177 |
| 32 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
-0.213 |
-0.050 |
| 33 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.205 |
-0.106 |
| 34 |
248484_at |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.204 |
0.024 |
| 35 |
245108_at |
AT2G41510
|
ATCKX1, CYTOKININ OXIDASE/DEHYDROGENASE 1, CKX1, cytokinin oxidase/dehydrogenase 1 |
-0.202 |
-0.113 |