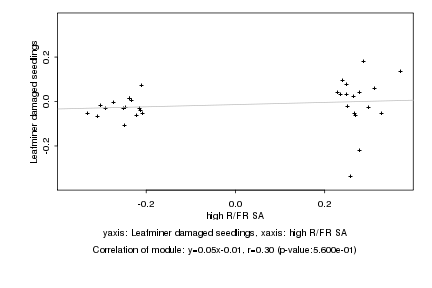
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
high R/FR SA |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.369 |
0.136 |
| 2 |
256672_at |
AT3G52310
|
ABCG27, ATP-binding cassette G27 |
0.326 |
-0.052 |
| 3 |
258340_at |
AT3G22770
|
[F-box associated ubiquitination effector family protein] |
0.310 |
0.060 |
| 4 |
245467_at |
AT4G16610
|
[C2H2-like zinc finger protein] |
0.298 |
-0.025 |
| 5 |
256147_at |
AT1G55080
|
MED9 |
0.286 |
0.182 |
| 6 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.278 |
0.043 |
| 7 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
0.276 |
-0.218 |
| 8 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
0.267 |
-0.062 |
| 9 |
255140_x_at |
AT4G08410
|
[Proline-rich extensin-like family protein] |
0.265 |
-0.052 |
| 10 |
259781_at |
AT1G29650
|
[non-LTR retrotransposon family (LINE), has a 1.5e-28 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
0.263 |
0.025 |
| 11 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
0.256 |
-0.336 |
| 12 |
245587_at |
AT4G15020
|
[hAT transposon superfamily] |
0.250 |
-0.019 |
| 13 |
252651_at |
AT3G44700
|
unknown |
0.248 |
0.033 |
| 14 |
247499_at |
AT5G61865
|
unknown |
0.247 |
0.077 |
| 15 |
260333_at |
AT1G70500
|
[Pectin lyase-like superfamily protein] |
0.239 |
0.097 |
| 16 |
264345_at |
AT1G11915
|
unknown |
0.234 |
0.033 |
| 17 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
0.228 |
0.043 |
| 18 |
256486_at |
AT1G31450
|
[Eukaryotic aspartyl protease family protein] |
-0.332 |
-0.053 |
| 19 |
263861_at |
AT2G04560
|
AtLpxB, LpxB, lipid X B |
-0.310 |
-0.067 |
| 20 |
256465_at |
AT1G32570
|
unknown |
-0.303 |
-0.015 |
| 21 |
261298_at |
AT1G48510
|
[Surfeit locus 1 cytochrome c oxidase biogenesis protein] |
-0.293 |
-0.028 |
| 22 |
264151_at |
AT1G02070
|
unknown |
-0.274 |
-0.004 |
| 23 |
263189_at |
AT1G36100
|
[myosin heavy chain-related] |
-0.251 |
-0.027 |
| 24 |
247802_at |
AT5G58580
|
ATL63, TOXICOS EN LEVADURA 63, TL63, TOXICOS EN LEVADURA 63 |
-0.249 |
-0.106 |
| 25 |
252253_at |
AT3G49300
|
[proline-rich family protein] |
-0.248 |
-0.026 |
| 26 |
252726_at |
AT3G43060
|
[pseudogene] |
-0.239 |
0.015 |
| 27 |
252319_at |
AT3G48710
|
[DEK domain-containing chromatin associated protein] |
-0.239 |
0.017 |
| 28 |
245091_at |
AT2G40910
|
[F-box and associated interaction domains-containing protein] |
-0.234 |
0.005 |
| 29 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.223 |
-0.062 |
| 30 |
265945_at |
AT2G19500
|
ATCKX2, CKX2, cytokinin oxidase 2 |
-0.217 |
-0.027 |
| 31 |
252755_at |
AT3G43530
|
unknown |
-0.213 |
-0.037 |
| 32 |
258722_at |
AT3G09590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.212 |
0.076 |
| 33 |
258639_at |
AT3G07820
|
[Pectin lyase-like superfamily protein] |
-0.209 |
-0.053 |