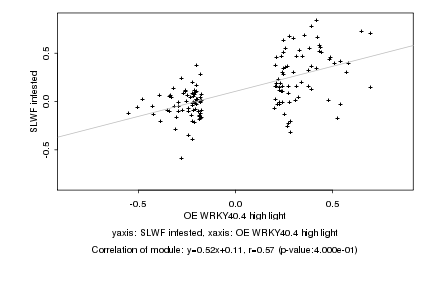
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
OE WRKY40.4 high light |
Log2 signal ratio
SLWF infested |
| 1 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.695 |
0.153 |
| 2 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.692 |
0.709 |
| 3 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.649 |
0.729 |
| 4 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.580 |
0.396 |
| 5 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.572 |
0.310 |
| 6 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.541 |
-0.029 |
| 7 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.540 |
0.415 |
| 8 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.523 |
-0.168 |
| 9 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.507 |
0.399 |
| 10 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.487 |
0.457 |
| 11 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.484 |
0.440 |
| 12 |
264998_at |
AT1G67330
|
unknown |
0.478 |
0.017 |
| 13 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.443 |
0.510 |
| 14 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.438 |
0.561 |
| 15 |
258203_at |
AT3G13950
|
unknown |
0.433 |
0.589 |
| 16 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.429 |
0.523 |
| 17 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.421 |
0.665 |
| 18 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.415 |
0.346 |
| 19 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.414 |
0.849 |
| 20 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.390 |
0.781 |
| 21 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.388 |
0.366 |
| 22 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.388 |
0.134 |
| 23 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.380 |
0.553 |
| 24 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.376 |
0.331 |
| 25 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.375 |
0.163 |
| 26 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.355 |
0.686 |
| 27 |
252345_at |
AT3G48640
|
unknown |
0.343 |
0.472 |
| 28 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.336 |
0.205 |
| 29 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.329 |
0.538 |
| 30 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
0.321 |
0.048 |
| 31 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.312 |
0.158 |
| 32 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.310 |
0.467 |
| 33 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
0.305 |
0.014 |
| 34 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
0.298 |
0.310 |
| 35 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.297 |
0.653 |
| 36 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
0.281 |
-0.319 |
| 37 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.281 |
-0.197 |
| 38 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.276 |
0.683 |
| 39 |
263126_at |
AT1G78460
|
[SOUL heme-binding family protein] |
0.274 |
-0.009 |
| 40 |
267412_at |
AT2G34940
|
BP80-3;2, binding protein of 80 kDa 3;2, VSR3;2, VACUOLAR SORTING RECEPTOR 3;2, VSR5, VACUOLAR SORTING RECEPTOR 5 |
0.273 |
0.159 |
| 41 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.271 |
-0.224 |
| 42 |
266824_at |
AT2G22800
|
HAT9 |
0.271 |
0.087 |
| 43 |
262085_at |
AT1G56060
|
unknown |
0.267 |
0.368 |
| 44 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.265 |
-0.256 |
| 45 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.257 |
0.554 |
| 46 |
248889_at |
AT5G46230
|
unknown |
0.257 |
0.356 |
| 47 |
260170_at |
AT1G71890
|
ATSUC5, SUCROSE-PROTON SYMPORTER 5, SUC5 |
0.251 |
-0.127 |
| 48 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.246 |
0.636 |
| 49 |
266017_at |
AT2G18690
|
unknown |
0.245 |
0.286 |
| 50 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.245 |
0.514 |
| 51 |
257798_at |
AT3G15950
|
NAI2 |
0.243 |
0.346 |
| 52 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.242 |
0.303 |
| 53 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
0.240 |
0.157 |
| 54 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.238 |
-0.004 |
| 55 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
0.238 |
0.114 |
| 56 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.236 |
0.475 |
| 57 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.235 |
0.113 |
| 58 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.228 |
0.193 |
| 59 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
0.227 |
0.152 |
| 60 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.225 |
-0.005 |
| 61 |
254710_at |
AT4G18050
|
ABCB9, ATP-binding cassette B9, PGP9, P-glycoprotein 9 |
0.223 |
-0.026 |
| 62 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.223 |
0.124 |
| 63 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.220 |
0.236 |
| 64 |
251645_at |
AT3G57790
|
[Pectin lyase-like superfamily protein] |
0.215 |
-0.024 |
| 65 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.210 |
0.165 |
| 66 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.209 |
0.466 |
| 67 |
260728_at |
AT1G48210
|
[Protein kinase superfamily protein] |
0.207 |
0.187 |
| 68 |
256603_at |
AT3G28270
|
unknown |
0.204 |
0.376 |
| 69 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.204 |
0.156 |
| 70 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.204 |
0.027 |
| 71 |
266618_at |
AT2G35480
|
unknown |
0.201 |
-0.063 |
| 72 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.554 |
-0.118 |
| 73 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.509 |
-0.056 |
| 74 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.483 |
0.023 |
| 75 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
-0.431 |
-0.051 |
| 76 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.424 |
-0.126 |
| 77 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
-0.394 |
0.065 |
| 78 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
-0.391 |
-0.202 |
| 79 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.353 |
-0.091 |
| 80 |
246108_at |
AT5G28630
|
[glycine-rich protein] |
-0.340 |
-0.094 |
| 81 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.340 |
0.062 |
| 82 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
-0.335 |
0.066 |
| 83 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
-0.333 |
0.051 |
| 84 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.321 |
0.138 |
| 85 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.317 |
-0.049 |
| 86 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.311 |
-0.287 |
| 87 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-0.305 |
-0.159 |
| 88 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.297 |
0.000 |
| 89 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
-0.296 |
-0.050 |
| 90 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.295 |
-0.094 |
| 91 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.282 |
-0.583 |
| 92 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.281 |
0.245 |
| 93 |
254201_at |
AT4G24130
|
unknown |
-0.277 |
-0.083 |
| 94 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.271 |
0.090 |
| 95 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.258 |
0.114 |
| 96 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.258 |
0.123 |
| 97 |
261315_at |
AT1G53170
|
ATERF-8, ATERF8, ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4, ERF8, ethylene response factor 8 |
-0.253 |
0.002 |
| 98 |
261069_at |
AT1G07410
|
ATRAB-A2B, ARABIDOPSIS RAB GTPASE HOMOLOG A2B, ATRABA2B, RAB GTPase homolog A2B, RAB-A2B, RAB GTPASE HOMOLOG A2B, RABA2b, RAB GTPase homolog A2B |
-0.252 |
0.071 |
| 99 |
250277_at |
AT5G12940
|
[Leucine-rich repeat (LRR) family protein] |
-0.247 |
-0.351 |
| 100 |
254034_at |
AT4G25960
|
ABCB2, ATP-binding cassette B2, PGP2, P-glycoprotein 2 |
-0.246 |
-0.063 |
| 101 |
263915_at |
AT2G36430
|
unknown |
-0.243 |
-0.095 |
| 102 |
260788_at |
AT1G06260
|
[Cysteine proteinases superfamily protein] |
-0.232 |
0.043 |
| 103 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
-0.230 |
-0.137 |
| 104 |
266485_at |
AT2G47630
|
[alpha/beta-Hydrolases superfamily protein] |
-0.226 |
-0.206 |
| 105 |
266968_at |
AT2G39360
|
[Protein kinase superfamily protein] |
-0.226 |
0.203 |
| 106 |
245776_at |
AT1G30260
|
unknown |
-0.225 |
-0.013 |
| 107 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.225 |
0.091 |
| 108 |
245406_at |
AT4G17160
|
ATRAB2B, ATRABB1A, RAB GTPase homolog B1A, RABB1a, RAB GTPase homolog B1A |
-0.224 |
-0.031 |
| 109 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.224 |
-0.388 |
| 110 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.218 |
-0.085 |
| 111 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
-0.218 |
0.052 |
| 112 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.218 |
-0.000 |
| 113 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.217 |
0.079 |
| 114 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.215 |
0.124 |
| 115 |
260820_at |
AT1G06840
|
[Leucine-rich repeat protein kinase family protein] |
-0.214 |
0.078 |
| 116 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.214 |
-0.209 |
| 117 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
-0.210 |
-0.019 |
| 118 |
245671_at |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
-0.209 |
0.099 |
| 119 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
-0.209 |
-0.080 |
| 120 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.208 |
0.003 |
| 121 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
-0.205 |
0.175 |
| 122 |
263690_at |
AT1G26960
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23 |
-0.204 |
0.022 |
| 123 |
256789_at |
AT3G13672
|
SINA2 |
-0.204 |
0.379 |
| 124 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.203 |
0.106 |
| 125 |
262286_at |
AT1G68585
|
unknown |
-0.201 |
0.021 |
| 126 |
250842_at |
AT5G04490
|
VTE5, vitamin E pathway gene 5 |
-0.201 |
-0.029 |
| 127 |
252037_at |
AT3G51920
|
ATCML9, CAM9, calmodulin 9, CML9, CALMODULIN LIKE PROTEIN 9 |
-0.200 |
0.029 |
| 128 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.195 |
-0.093 |
| 129 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.187 |
-0.154 |
| 130 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.187 |
-0.138 |
| 131 |
256671_at |
AT3G52290
|
IQD3, IQ-domain 3 |
-0.187 |
-0.177 |
| 132 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
-0.185 |
0.042 |
| 133 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.185 |
-0.119 |
| 134 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.184 |
-0.166 |
| 135 |
259418_at |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
-0.183 |
0.280 |
| 136 |
265075_at |
AT1G55450
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.183 |
-0.006 |
| 137 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
-0.182 |
0.006 |
| 138 |
246233_at |
AT4G36550
|
[ARM repeat superfamily protein] |
-0.180 |
0.017 |
| 139 |
260876_at |
AT1G21460
|
AtSWEET1, SWEET1 |
-0.178 |
-0.083 |
| 140 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.178 |
-0.163 |
| 141 |
247455_at |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
-0.177 |
0.078 |