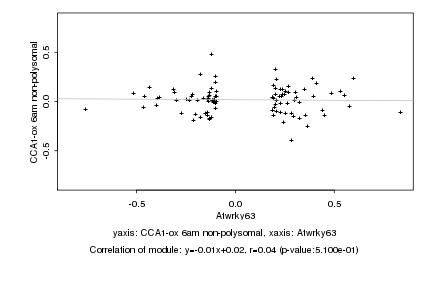
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Atwrky63 |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
249598_at |
AT5G37970
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.833 |
-0.109 |
| 2 |
256636_at |
AT3G12000
|
[S-locus related protein SLR1, putative (S1)] |
0.593 |
0.234 |
| 3 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
0.574 |
-0.041 |
| 4 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
0.551 |
0.066 |
| 5 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.529 |
0.102 |
| 6 |
256335_at |
AT1G72110
|
[O-acyltransferase (WSD1-like) family protein] |
0.485 |
0.084 |
| 7 |
264151_at |
AT1G02070
|
unknown |
0.449 |
-0.141 |
| 8 |
245571_at |
AT4G14695
|
[Uncharacterised protein family (UPF0041)] |
0.438 |
-0.083 |
| 9 |
259526_at |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.405 |
0.193 |
| 10 |
258058_at |
AT3G28980
|
unknown |
0.390 |
0.058 |
| 11 |
256588_at |
AT3G28790
|
unknown |
0.385 |
0.237 |
| 12 |
255912_at |
AT1G66960
|
LUP5 |
0.364 |
-0.247 |
| 13 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
0.350 |
-0.132 |
| 14 |
259549_at |
AT1G35290
|
ALT1, acyl-lipid thioesterase 1 |
0.347 |
0.127 |
| 15 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.322 |
-0.170 |
| 16 |
265261_at |
AT2G42990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.321 |
-0.008 |
| 17 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
0.305 |
0.044 |
| 18 |
266549_at |
AT2G35150
|
EXL7, EXORDIUM LIKE 7 |
0.300 |
0.101 |
| 19 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
0.294 |
0.015 |
| 20 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.289 |
-0.148 |
| 21 |
251448_at |
AT3G59845
|
[Zinc-binding dehydrogenase family protein] |
0.282 |
-0.393 |
| 22 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
0.280 |
-0.119 |
| 23 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
0.266 |
0.153 |
| 24 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.265 |
0.098 |
| 25 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.260 |
-0.019 |
| 26 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
0.251 |
-0.112 |
| 27 |
247882_at |
AT5G57785
|
unknown |
0.250 |
0.103 |
| 28 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
0.245 |
0.076 |
| 29 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.242 |
-0.205 |
| 30 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.235 |
0.075 |
| 31 |
258962_at |
AT3G10570
|
CYP77A6, cytochrome P450, family 77, subfamily A, polypeptide 6 |
0.234 |
0.123 |
| 32 |
257815_at |
AT3G25130
|
unknown |
0.231 |
0.052 |
| 33 |
250631_at |
AT5G07430
|
[Pectin lyase-like superfamily protein] |
0.227 |
-0.108 |
| 34 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.223 |
0.131 |
| 35 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
0.223 |
-0.015 |
| 36 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.222 |
0.053 |
| 37 |
254595_at |
AT4G18960
|
AG, AGAMOUS |
0.206 |
0.225 |
| 38 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.205 |
-0.096 |
| 39 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.205 |
0.018 |
| 40 |
265536_at |
AT2G15880
|
[Leucine-rich repeat (LRR) family protein] |
0.200 |
0.330 |
| 41 |
264137_at |
AT1G78960
|
ATLUP2, lupeol synthase 2, LUP2, lupeol synthase 2 |
0.199 |
-0.025 |
| 42 |
257679_at |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
0.198 |
0.136 |
| 43 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.198 |
0.081 |
| 44 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.193 |
-0.052 |
| 45 |
256145_at |
AT1G48750
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.191 |
0.033 |
| 46 |
248924_at |
AT5G45960
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.189 |
-0.134 |
| 47 |
246913_at |
AT5G25830
|
GATA12, GATA transcription factor 12 |
0.189 |
0.164 |
| 48 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
0.186 |
0.047 |
| 49 |
260226_at |
AT1G74660
|
MIF1, mini zinc finger 1 |
0.185 |
-0.083 |
| 50 |
251903_at |
AT3G54120
|
[Reticulon family protein] |
0.183 |
-0.087 |
| 51 |
250872_at |
AT5G03960
|
IQD12, IQ-domain 12 |
0.183 |
0.042 |
| 52 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.760 |
-0.073 |
| 53 |
259385_at |
AT1G13470
|
unknown |
-0.517 |
0.085 |
| 54 |
258897_at |
AT3G05730
|
[LOCATED IN: endomembrane system] |
-0.468 |
-0.056 |
| 55 |
263851_at |
AT2G04460
|
unknown |
-0.461 |
0.059 |
| 56 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.436 |
0.148 |
| 57 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.402 |
-0.031 |
| 58 |
246001_at |
AT5G20790
|
unknown |
-0.398 |
0.035 |
| 59 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.387 |
0.051 |
| 60 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.316 |
0.123 |
| 61 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.311 |
0.098 |
| 62 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
-0.300 |
0.014 |
| 63 |
264635_at |
AT1G65500
|
unknown |
-0.275 |
-0.118 |
| 64 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.249 |
0.027 |
| 65 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
-0.233 |
0.013 |
| 66 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.224 |
0.056 |
| 67 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-0.219 |
0.080 |
| 68 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.214 |
-0.184 |
| 69 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.207 |
-0.130 |
| 70 |
254112_at |
AT4G24970
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
-0.194 |
0.012 |
| 71 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.180 |
0.283 |
| 72 |
260734_at |
AT1G17600
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.177 |
-0.157 |
| 73 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.162 |
0.039 |
| 74 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.156 |
-0.114 |
| 75 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.144 |
0.022 |
| 76 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
-0.142 |
-0.105 |
| 77 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
-0.142 |
-0.134 |
| 78 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.141 |
0.004 |
| 79 |
264660_at |
AT1G09940
|
HEMA2 |
-0.141 |
0.060 |
| 80 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.138 |
0.036 |
| 81 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.135 |
-0.166 |
| 82 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.134 |
-0.175 |
| 83 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
-0.134 |
0.099 |
| 84 |
257765_at |
AT3G23020
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.132 |
0.067 |
| 85 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.125 |
0.133 |
| 86 |
244995_at |
ATCG00150
|
ATPI |
-0.124 |
0.482 |
| 87 |
262719_at |
AT1G43590
|
unknown |
-0.122 |
-0.156 |
| 88 |
257322_at |
ATMG01180
|
ORF111B |
-0.121 |
0.020 |
| 89 |
265032_at |
AT1G61580
|
ARP2, ARABIDOPSIS RIBOSOMAL PROTEIN 2, RPL3B, R-protein L3 B |
-0.119 |
0.015 |
| 90 |
257796_at |
AT3G15930
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.114 |
0.016 |
| 91 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.112 |
-0.004 |
| 92 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
-0.111 |
0.010 |
| 93 |
258190_at |
AT3G29080
|
unknown |
-0.106 |
0.022 |
| 94 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.104 |
0.255 |
| 95 |
250146_at |
AT5G14660
|
ATDEF2, DEF2, PDF1B, peptide deformylase 1B |
-0.103 |
0.202 |
| 96 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.103 |
-0.064 |
| 97 |
255904_at |
AT1G17860
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.102 |
0.059 |
| 98 |
267142_at |
AT2G38290
|
AMT2, ammonium transporter 2, AMT2;1, AMMONIUM TRANSPORTER 2;1, ATAMT2, ammonium transporter 2 |
-0.101 |
-0.018 |
| 99 |
267219_at |
AT2G02590
|
unknown |
-0.097 |
0.102 |
| 100 |
257636_at |
AT3G26200
|
CYP71B22, cytochrome P450, family 71, subfamily B, polypeptide 22 |
-0.097 |
0.055 |
| 101 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.097 |
0.005 |