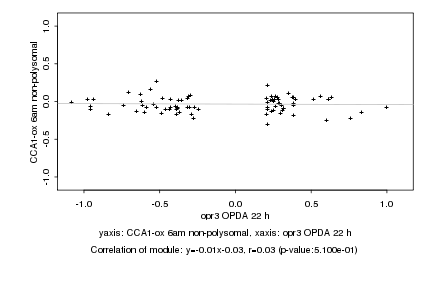
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
opr3 OPDA 22 h |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
249428_at |
AT5G39870
|
unknown |
0.997 |
-0.077 |
| 2 |
267531_at |
AT2G41860
|
CPK14, calcium-dependent protein kinase 14 |
0.826 |
-0.140 |
| 3 |
255003_at |
AT4G09950
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.755 |
-0.212 |
| 4 |
258129_at |
AT3G24620
|
ATROPGEF8, ROPGEF8, ROP (rho of plants) guanine nucleotide exchange factor 8 |
0.629 |
0.056 |
| 5 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.609 |
0.033 |
| 6 |
248711_at |
AT5G48270
|
unknown |
0.608 |
0.038 |
| 7 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.600 |
-0.251 |
| 8 |
255831_at |
AT2G33350
|
[CCT motif family protein] |
0.557 |
0.069 |
| 9 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.512 |
0.040 |
| 10 |
251635_at |
AT3G57510
|
ADPG1 |
0.394 |
0.034 |
| 11 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.383 |
-0.025 |
| 12 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.383 |
-0.185 |
| 13 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.382 |
0.058 |
| 14 |
260012_at |
AT1G67865
|
unknown |
0.377 |
-0.049 |
| 15 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.375 |
0.057 |
| 16 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.345 |
0.112 |
| 17 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
0.314 |
-0.089 |
| 18 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.313 |
-0.083 |
| 19 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.306 |
-0.106 |
| 20 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.300 |
-0.050 |
| 21 |
257670_at |
AT3G20340
|
unknown |
0.294 |
-0.153 |
| 22 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
0.288 |
-0.023 |
| 23 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.281 |
0.034 |
| 24 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.280 |
-0.021 |
| 25 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.273 |
0.054 |
| 26 |
250881_at |
AT5G04080
|
unknown |
0.264 |
0.074 |
| 27 |
245635_at |
AT1G25250
|
AtIDD16, indeterminate(ID)-domain 16, IDD16, indeterminate(ID)-domain 16 |
0.260 |
-0.058 |
| 28 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.256 |
0.032 |
| 29 |
249136_at |
AT5G43180
|
unknown |
0.253 |
0.034 |
| 30 |
264127_at |
AT1G79250
|
AGC1.7, AGC kinase 1.7 |
0.251 |
-0.113 |
| 31 |
262936_at |
AT1G79400
|
ATCHX2, cation/H+ exchanger 2, CHX2, cation/H+ exchanger 2 |
0.245 |
0.003 |
| 32 |
265732_at |
AT2G01300
|
unknown |
0.240 |
0.030 |
| 33 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.238 |
-0.122 |
| 34 |
257196_at |
AT3G23790
|
AAE16, acyl activating enzyme 16 |
0.233 |
0.076 |
| 35 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
0.227 |
0.015 |
| 36 |
250449_at |
AT5G10830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.211 |
-0.066 |
| 37 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.211 |
0.215 |
| 38 |
246250_at |
AT4G36880
|
CP1, cysteine proteinase1, RDL1, RD21A-LIKE PROTEASE1 |
0.211 |
-0.095 |
| 39 |
264430_at |
AT1G61680
|
ATTPS14, TERPENE SYNTHASE 14, TPS14, terpene synthase 14 |
0.210 |
-0.294 |
| 40 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
0.208 |
-0.002 |
| 41 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.203 |
0.046 |
| 42 |
267358_at |
AT2G39890
|
ATPROT1, PROLINE TRANSPORTER 1, PROT1, proline transporter 1 |
0.202 |
-0.159 |
| 43 |
262697_at |
AT1G75940
|
ATA27, BGLU20, BETA GLUCOSIDASE 20 |
-1.087 |
-0.010 |
| 44 |
257102_at |
AT3G25050
|
XTH3, xyloglucan endotransglucosylase/hydrolase 3 |
-0.977 |
0.039 |
| 45 |
262674_at |
AT1G75910
|
EXL4, extracellular lipase 4 |
-0.960 |
-0.100 |
| 46 |
253087_at |
AT4G36350
|
ATPAP25, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 25, PAP25, purple acid phosphatase 25 |
-0.958 |
-0.058 |
| 47 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
-0.941 |
0.036 |
| 48 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.839 |
-0.161 |
| 49 |
256481_at |
AT1G33430
|
[Galactosyltransferase family protein] |
-0.740 |
-0.051 |
| 50 |
245652_at |
AT4G13960
|
[F-box/RNI-like superfamily protein] |
-0.708 |
0.124 |
| 51 |
248872_at |
AT5G46795
|
MSP2, microspore-specific promoter 2 |
-0.654 |
-0.124 |
| 52 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.633 |
0.106 |
| 53 |
256059_at |
AT1G06990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.622 |
0.012 |
| 54 |
257399_at |
AT1G23690
|
unknown |
-0.617 |
-0.041 |
| 55 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
-0.603 |
-0.133 |
| 56 |
260788_at |
AT1G06260
|
[Cysteine proteinases superfamily protein] |
-0.589 |
-0.075 |
| 57 |
252005_at |
AT3G52810
|
ATPAP21, PURPLE ACID PHOSPHATASE 21, PAP21, purple acid phosphatase 21 |
-0.566 |
0.163 |
| 58 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
-0.544 |
-0.031 |
| 59 |
255806_at |
AT4G10260
|
[pfkB-like carbohydrate kinase family protein] |
-0.523 |
0.269 |
| 60 |
245785_at |
AT1G32180
|
ATCSLD6, cellulose synthase-like D6, CSLD6, CELLULOSE SYNTHASE LIKE D6, CSLD6, cellulose synthase-like D6 |
-0.523 |
-0.066 |
| 61 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
-0.493 |
-0.157 |
| 62 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.487 |
0.047 |
| 63 |
253862_at |
AT4G27330
|
NZZ, NOZZLE, SPL, SPOROCYTELESS |
-0.463 |
-0.093 |
| 64 |
260228_at |
AT1G74540
|
CYP98A8, cytochrome P450, family 98, subfamily A, polypeptide 8 |
-0.436 |
-0.101 |
| 65 |
258767_at |
AT3G10890
|
[Glycosyl hydrolase superfamily protein] |
-0.434 |
-0.074 |
| 66 |
254494_at |
AT4G20050
|
QRT3, QUARTET 3 |
-0.433 |
0.038 |
| 67 |
255337_at |
AT4G04450
|
AtWRKY42, WRKY42 |
-0.402 |
-0.059 |
| 68 |
264431_at |
AT1G61700
|
[RNA polymerases N / 8 kDa subunit] |
-0.395 |
-0.102 |
| 69 |
256307_at |
AT1G30350
|
[Pectin lyase-like superfamily protein] |
-0.391 |
-0.170 |
| 70 |
262675_at |
AT1G75930
|
EXL6, extracellular lipase 6 |
-0.387 |
-0.069 |
| 71 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.387 |
-0.082 |
| 72 |
250636_at |
AT5G07520
|
ATGRP-8, ATGRP18, GRP18, glycine-rich protein 18 |
-0.378 |
0.022 |
| 73 |
250251_at |
AT5G13670
|
UMAMIT15, Usually multiple acids move in and out Transporters 15 |
-0.375 |
-0.144 |
| 74 |
258392_at |
AT3G15400
|
ATA20, anther 20 |
-0.359 |
0.015 |
| 75 |
261392_at |
AT1G79780
|
[Uncharacterised protein family (UPF0497)] |
-0.321 |
-0.070 |
| 76 |
261883_at |
AT1G80870
|
[Protein kinase superfamily protein] |
-0.320 |
0.047 |
| 77 |
250532_at |
AT5G08540
|
unknown |
-0.313 |
0.076 |
| 78 |
250638_at |
AT5G07540
|
ATGRP-6, ATGRP16, GRP16, glycine-rich protein 16 |
-0.308 |
-0.079 |
| 79 |
256381_at |
AT1G66850
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.299 |
0.089 |
| 80 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.294 |
-0.160 |
| 81 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-0.277 |
-0.213 |
| 82 |
260563_at |
AT2G43840
|
UGT74F1, UDP-glycosyltransferase 74 F1 |
-0.275 |
-0.078 |
| 83 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
-0.244 |
-0.104 |