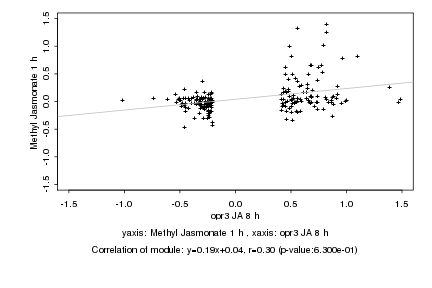
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
opr3 JA 8 h |
Log2 signal ratio
Methyl Jasmonate 1 h |
| 1 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
1.483 |
0.049 |
| 2 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
1.466 |
-0.009 |
| 3 |
249349_at |
AT5G40350
|
AtMYB24, myb domain protein 24, MYB24, myb domain protein 24 |
1.381 |
0.265 |
| 4 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.096 |
0.822 |
| 5 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
0.997 |
0.035 |
| 6 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.983 |
0.012 |
| 7 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.964 |
0.781 |
| 8 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.953 |
-0.027 |
| 9 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.918 |
0.139 |
| 10 |
250264_at |
AT5G12890
|
[UDP-Glycosyltransferase superfamily protein] |
0.915 |
0.276 |
| 11 |
258631_at |
AT3G07970
|
QRT2, QUARTET 2 |
0.909 |
0.060 |
| 12 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.878 |
0.096 |
| 13 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
0.876 |
-0.053 |
| 14 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.869 |
0.085 |
| 15 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.869 |
-0.259 |
| 16 |
260621_at |
AT1G08065
|
ACA5, alpha carbonic anhydrase 5, ATACA5, ALPHA CARBONIC ANHYDRASE 5 |
0.862 |
0.020 |
| 17 |
254629_at |
AT4G18425
|
unknown |
0.857 |
-0.000 |
| 18 |
256757_at |
AT3G25620
|
ABCG21, ATP-binding cassette G21 |
0.831 |
-0.002 |
| 19 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.817 |
1.401 |
| 20 |
245443_at |
AT4G16730
|
TPS02, terpene synthase 02 |
0.817 |
0.049 |
| 21 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.815 |
1.263 |
| 22 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.804 |
0.088 |
| 23 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.791 |
-0.144 |
| 24 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.787 |
1.024 |
| 25 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.782 |
0.539 |
| 26 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.767 |
0.668 |
| 27 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.747 |
0.619 |
| 28 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.739 |
0.386 |
| 29 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.736 |
0.096 |
| 30 |
250535_at |
AT5G08480
|
[VQ motif-containing protein] |
0.736 |
-0.017 |
| 31 |
252010_at |
AT3G52740
|
unknown |
0.733 |
-0.132 |
| 32 |
252920_at |
AT4G39000
|
AtGH9B17, glycosyl hydrolase 9B17, GH9B17, glycosyl hydrolase 9B17 |
0.722 |
0.000 |
| 33 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.712 |
-0.085 |
| 34 |
247193_at |
AT5G65380
|
[MATE efflux family protein] |
0.691 |
0.207 |
| 35 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
0.683 |
-0.008 |
| 36 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.678 |
0.667 |
| 37 |
245542_at |
AT4G15250
|
BBX9, B-box domain protein 9 |
0.677 |
0.077 |
| 38 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.677 |
0.105 |
| 39 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.674 |
0.667 |
| 40 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
0.670 |
-0.029 |
| 41 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.668 |
0.101 |
| 42 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.664 |
-0.007 |
| 43 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.651 |
0.023 |
| 44 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
0.650 |
0.495 |
| 45 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.644 |
0.244 |
| 46 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.643 |
0.314 |
| 47 |
260885_at |
AT1G29230
|
ATCIPK18, ATWL1, WPL4-LIKE 1, CIPK18, CBL-interacting protein kinase 18, SnRK3.20, SNF1-RELATED PROTEIN KINASE 3.20, WL1, WPL4-LIKE 1 |
0.638 |
0.181 |
| 48 |
251635_at |
AT3G57510
|
ADPG1 |
0.636 |
0.060 |
| 49 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.609 |
0.175 |
| 50 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
0.591 |
0.018 |
| 51 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.590 |
0.304 |
| 52 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.585 |
-0.170 |
| 53 |
265059_at |
AT1G52080
|
AR791 |
0.584 |
0.040 |
| 54 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.573 |
0.277 |
| 55 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.554 |
1.325 |
| 56 |
246256_at |
AT4G36770
|
[UDP-Glycosyltransferase superfamily protein] |
0.554 |
0.008 |
| 57 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
0.553 |
-0.192 |
| 58 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
0.553 |
0.373 |
| 59 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
0.552 |
0.065 |
| 60 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.545 |
-0.002 |
| 61 |
259735_at |
AT1G64405
|
unknown |
0.542 |
-0.180 |
| 62 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.537 |
0.431 |
| 63 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.532 |
-0.016 |
| 64 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
0.531 |
-0.031 |
| 65 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.531 |
0.000 |
| 66 |
262795_at |
AT1G13130
|
[Cellulase (glycosyl hydrolase family 5) protein] |
0.528 |
-0.025 |
| 67 |
258201_at |
AT3G13910
|
unknown |
0.520 |
0.111 |
| 68 |
259980_at |
AT1G76520
|
[Auxin efflux carrier family protein] |
0.515 |
0.056 |
| 69 |
259596_at |
AT1G28130
|
GH3.17 |
0.511 |
-0.326 |
| 70 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.508 |
0.496 |
| 71 |
248643_at |
AT5G49130
|
[MATE efflux family protein] |
0.508 |
-0.021 |
| 72 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
0.508 |
0.039 |
| 73 |
264588_at |
AT2G17730
|
NIP2, NEP-interacting protein 2 |
0.504 |
-0.189 |
| 74 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.498 |
0.036 |
| 75 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.497 |
0.828 |
| 76 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.497 |
0.043 |
| 77 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
0.496 |
-0.088 |
| 78 |
245682_at |
AT5G08750
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.483 |
-0.114 |
| 79 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.482 |
0.099 |
| 80 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.479 |
0.998 |
| 81 |
260387_at |
AT1G74100
|
ATSOT16, SULFOTRANSFERASE 16, ATST5A, ARABIDOPSIS SULFOTRANSFERASE 5A, CORI-7, CORONATINE INDUCED-7, SOT16, sulfotransferase 16 |
0.474 |
0.233 |
| 82 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
0.471 |
0.190 |
| 83 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
0.470 |
0.412 |
| 84 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.461 |
0.007 |
| 85 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.455 |
-0.167 |
| 86 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.455 |
0.170 |
| 87 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.455 |
-0.325 |
| 88 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
0.447 |
-0.083 |
| 89 |
257409_at |
AT2G17470
|
ALMT6, ALuminium activated Malate Transporter 6, AtALMT6 |
0.445 |
0.188 |
| 90 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.445 |
0.496 |
| 91 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.444 |
0.627 |
| 92 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
0.442 |
-0.002 |
| 93 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.431 |
0.168 |
| 94 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.429 |
0.044 |
| 95 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
0.427 |
0.236 |
| 96 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
0.425 |
-0.063 |
| 97 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.417 |
-0.077 |
| 98 |
261772_at |
AT1G76240
|
unknown |
0.415 |
-0.021 |
| 99 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.413 |
-0.151 |
| 100 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.412 |
0.038 |
| 101 |
245439_at |
AT4G16670
|
unknown |
0.411 |
0.140 |
| 102 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-1.026 |
0.022 |
| 103 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
-0.739 |
0.063 |
| 104 |
245652_at |
AT4G13960
|
[F-box/RNI-like superfamily protein] |
-0.617 |
0.045 |
| 105 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.547 |
0.135 |
| 106 |
249013_at |
AT5G44700
|
EDA23, EMBRYO SAC DEVELOPMENT ARREST 23, GSO2, GASSHO 2 |
-0.531 |
-0.007 |
| 107 |
253862_at |
AT4G27330
|
NZZ, NOZZLE, SPL, SPOROCYTELESS |
-0.514 |
0.042 |
| 108 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.508 |
0.067 |
| 109 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.508 |
0.033 |
| 110 |
261392_at |
AT1G79780
|
[Uncharacterised protein family (UPF0497)] |
-0.499 |
0.028 |
| 111 |
262452_at |
AT1G11210
|
unknown |
-0.493 |
-0.075 |
| 112 |
247535_at |
AT5G61620
|
[myb-like transcription factor family protein] |
-0.492 |
-0.041 |
| 113 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.479 |
-0.025 |
| 114 |
249134_at |
AT5G43150
|
unknown |
-0.472 |
0.060 |
| 115 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.467 |
0.230 |
| 116 |
248028_at |
AT5G55620
|
unknown |
-0.462 |
-0.456 |
| 117 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.461 |
-0.068 |
| 118 |
265468_at |
AT2G37210
|
LOG3, LONELY GUY 3 |
-0.456 |
0.059 |
| 119 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.454 |
-0.165 |
| 120 |
264580_at |
AT1G05340
|
unknown |
-0.454 |
-0.100 |
| 121 |
257779_at |
AT3G26940
|
CDG1, CONSTITUTIVE DIFFERENTIAL GROWTH 1 |
-0.450 |
-0.021 |
| 122 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
-0.428 |
-0.116 |
| 123 |
245436_at |
AT4G16620
|
UMAMIT8, Usually multiple acids move in and out Transporters 8 |
-0.425 |
0.062 |
| 124 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.416 |
0.026 |
| 125 |
267226_at |
AT2G44010
|
unknown |
-0.398 |
-0.029 |
| 126 |
260264_at |
AT1G68500
|
unknown |
-0.397 |
0.064 |
| 127 |
250251_at |
AT5G13670
|
UMAMIT15, Usually multiple acids move in and out Transporters 15 |
-0.378 |
0.077 |
| 128 |
250937_at |
AT5G03230
|
unknown |
-0.377 |
-0.306 |
| 129 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
-0.374 |
-0.030 |
| 130 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.359 |
0.180 |
| 131 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.355 |
0.037 |
| 132 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.352 |
-0.049 |
| 133 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.351 |
0.023 |
| 134 |
252408_at |
AT3G47600
|
ATMYB94, myb domain protein 94, ATMYBCP70, MYB94, myb domain protein 94 |
-0.347 |
0.048 |
| 135 |
260563_at |
AT2G43840
|
UGT74F1, UDP-glycosyltransferase 74 F1 |
-0.346 |
0.092 |
| 136 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-0.339 |
-0.021 |
| 137 |
256674_at |
AT3G52360
|
unknown |
-0.332 |
-0.204 |
| 138 |
253672_at |
AT4G29820
|
ATCFIM-25, ARABIDOPSIS THALIANA HOMOLOG OF CFIM-25, CFIM-25, homolog of CFIM-25 |
-0.324 |
0.038 |
| 139 |
252930_at |
AT4G39010
|
AtGH9B18, glycosyl hydrolase 9B18, GH9B18, glycosyl hydrolase 9B18 |
-0.324 |
-0.001 |
| 140 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.321 |
-0.044 |
| 141 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.321 |
-0.113 |
| 142 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.319 |
0.041 |
| 143 |
256429_at |
AT3G11040
|
AtENGase85B, ENGase85B, Endo-beta-N-acetyglucosaminidase 85B |
-0.319 |
0.008 |
| 144 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.318 |
-0.062 |
| 145 |
256114_at |
AT1G16850
|
unknown |
-0.312 |
0.077 |
| 146 |
260559_at |
AT2G43860
|
[Pectin lyase-like superfamily protein] |
-0.311 |
-0.041 |
| 147 |
246532_at |
AT5G15870
|
[glycosyl hydrolase family 81 protein] |
-0.311 |
-0.076 |
| 148 |
252730_at |
AT3G43110
|
unknown |
-0.304 |
0.365 |
| 149 |
252073_at |
AT3G51750
|
unknown |
-0.300 |
0.051 |
| 150 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.290 |
-0.119 |
| 151 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.289 |
-0.108 |
| 152 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
-0.289 |
0.062 |
| 153 |
264790_at |
AT2G17820
|
AHK1, ATHK1, histidine kinase 1, HK1, histidine kinase 1 |
-0.289 |
-0.292 |
| 154 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.288 |
0.069 |
| 155 |
263544_at |
AT2G21590
|
APL4 |
-0.288 |
0.060 |
| 156 |
260345_at |
AT1G69270
|
AtRPK1, RPK1, receptor-like protein kinase 1 |
-0.287 |
-0.142 |
| 157 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.285 |
0.174 |
| 158 |
267036_at |
AT2G38465
|
unknown |
-0.281 |
-0.041 |
| 159 |
260490_at |
AT1G51500
|
ABCG12, ATP-binding cassette G12, AtABCG12, ATWBC12, ARABIDOPSIS THALIANA WHITE-BROWN COMPLEX 12, CER5, ECERIFERUM 5, D3, WBC12, WHITE-BROWN COMPLEX 12 |
-0.281 |
-0.079 |
| 160 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
-0.279 |
-0.046 |
| 161 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
-0.278 |
0.015 |
| 162 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
-0.276 |
0.075 |
| 163 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
-0.268 |
-0.040 |
| 164 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.264 |
-0.038 |
| 165 |
262436_at |
AT1G47610
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.263 |
-0.020 |
| 166 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
-0.262 |
-0.100 |
| 167 |
245758_at |
AT1G32240
|
KAN2, KANADI 2 |
-0.259 |
-0.066 |
| 168 |
260797_at |
AT1G78390
|
ATNCED9, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9, NCED9, nine-cis-epoxycarotenoid dioxygenase 9 |
-0.256 |
0.008 |
| 169 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.254 |
-0.297 |
| 170 |
261062_at |
AT1G07530
|
ATGRAS2, ARABIDOPSIS THALIANA GRAS (GAI, RGA, SCR) 2, GRAS2, GRAS (GAI, RGA, SCR) 2, SCL14, SCARECROW-like 14 |
-0.251 |
-0.073 |
| 171 |
261448_at |
AT1G21140
|
[Vacuolar iron transporter (VIT) family protein] |
-0.250 |
0.135 |
| 172 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.250 |
-0.159 |
| 173 |
263183_at |
AT1G05570
|
ATGSL06, ATGSL6, CALS1, callose synthase 1, GSL06, GSL6, GLUCAN SYNTHASE-LIKE 6 |
-0.249 |
-0.016 |
| 174 |
245602_at |
AT4G14270
|
unknown |
-0.249 |
-0.165 |
| 175 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-0.249 |
-0.238 |
| 176 |
265480_at |
AT2G15970
|
ATCOR413-PM1, ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1, ATCYP19, CYCLOPHILLIN 19, COR413-PM1, cold regulated 413 plasma membrane 1, FL3-5A3, WCOR413, WCOR413-LIKE |
-0.248 |
-0.013 |
| 177 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
-0.247 |
-0.043 |
| 178 |
261089_at |
AT1G07570
|
APK1, APK1A |
-0.245 |
-0.201 |
| 179 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.244 |
-0.164 |
| 180 |
266126_at |
AT2G45040
|
[Matrixin family protein] |
-0.244 |
0.065 |
| 181 |
263265_at |
AT2G38820
|
unknown |
-0.241 |
-0.203 |
| 182 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.240 |
-0.288 |
| 183 |
250938_at |
AT5G03180
|
[RING/U-box superfamily protein] |
-0.240 |
-0.050 |
| 184 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
-0.239 |
0.040 |
| 185 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
-0.238 |
-0.033 |
| 186 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
-0.231 |
-0.030 |
| 187 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.230 |
-0.058 |
| 188 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
-0.229 |
0.135 |
| 189 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.227 |
0.068 |
| 190 |
258374_at |
AT3G14360
|
[alpha/beta-Hydrolases superfamily protein] |
-0.225 |
0.005 |
| 191 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.225 |
-0.171 |
| 192 |
266006_at |
AT2G37360
|
ABCG2, ATP-binding cassette G2 |
-0.223 |
0.155 |
| 193 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.223 |
-0.057 |
| 194 |
253188_at |
AT4G35300
|
TMT2, tonoplast monosaccharide transporter2 |
-0.223 |
-0.010 |
| 195 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.218 |
-0.108 |
| 196 |
257232_at |
AT3G16500
|
IAA26, indole-3-acetic acid inducible 26, PAP1, phytochrome-associated protein 1 |
-0.218 |
0.171 |
| 197 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
-0.217 |
0.058 |
| 198 |
248558_at |
AT5G49990
|
[Xanthine/uracil permease family protein] |
-0.216 |
-0.168 |
| 199 |
247585_at |
AT5G60680
|
unknown |
-0.213 |
-0.430 |
| 200 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.212 |
0.030 |
| 201 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.211 |
-0.369 |
| 202 |
249922_at |
AT5G19140
|
AILP1, ATAILP1 |
-0.211 |
-0.000 |