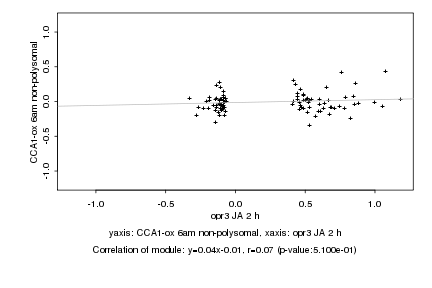
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
opr3 JA 2 h |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
1.180 |
0.033 |
| 2 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.073 |
0.439 |
| 3 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.048 |
-0.070 |
| 4 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.995 |
-0.007 |
| 5 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.877 |
-0.024 |
| 6 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.858 |
0.266 |
| 7 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
0.847 |
-0.030 |
| 8 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.846 |
0.085 |
| 9 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.823 |
-0.237 |
| 10 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.788 |
0.068 |
| 11 |
250264_at |
AT5G12890
|
[UDP-Glycosyltransferase superfamily protein] |
0.777 |
-0.096 |
| 12 |
266456_at |
AT2G22770
|
NAI1 |
0.757 |
0.421 |
| 13 |
249349_at |
AT5G40350
|
AtMYB24, myb domain protein 24, MYB24, myb domain protein 24 |
0.740 |
-0.067 |
| 14 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
0.705 |
-0.087 |
| 15 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.686 |
-0.076 |
| 16 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.678 |
-0.083 |
| 17 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
0.668 |
-0.178 |
| 18 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.665 |
0.028 |
| 19 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
0.652 |
0.210 |
| 20 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
0.633 |
-0.025 |
| 21 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.628 |
-0.089 |
| 22 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
0.607 |
-0.130 |
| 23 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.601 |
0.040 |
| 24 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.596 |
-0.038 |
| 25 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.592 |
-0.136 |
| 26 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.569 |
-0.213 |
| 27 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.538 |
0.038 |
| 28 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.529 |
-0.076 |
| 29 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.528 |
0.022 |
| 30 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.528 |
-0.344 |
| 31 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
0.522 |
-0.000 |
| 32 |
248643_at |
AT5G49130
|
[MATE efflux family protein] |
0.514 |
0.057 |
| 33 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.512 |
-0.149 |
| 34 |
250960_at |
AT5G02940
|
unknown |
0.504 |
0.034 |
| 35 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.496 |
0.020 |
| 36 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.482 |
0.112 |
| 37 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
0.482 |
0.022 |
| 38 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.482 |
0.098 |
| 39 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.482 |
-0.095 |
| 40 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.470 |
-0.081 |
| 41 |
255527_at |
AT4G02360
|
unknown |
0.464 |
0.179 |
| 42 |
259518_at |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
0.461 |
-0.050 |
| 43 |
264873_at |
AT1G24100
|
UGT74B1, UDP-glucosyl transferase 74B1 |
0.456 |
-0.112 |
| 44 |
249025_at |
AT5G44720
|
[Molybdenum cofactor sulfurase family protein] |
0.452 |
-0.014 |
| 45 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.443 |
0.123 |
| 46 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.439 |
0.080 |
| 47 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.438 |
0.038 |
| 48 |
264289_at |
AT1G61890
|
[MATE efflux family protein] |
0.427 |
0.249 |
| 49 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.416 |
0.316 |
| 50 |
260387_at |
AT1G74100
|
ATSOT16, SULFOTRANSFERASE 16, ATST5A, ARABIDOPSIS SULFOTRANSFERASE 5A, CORI-7, CORONATINE INDUCED-7, SOT16, sulfotransferase 16 |
0.413 |
0.006 |
| 51 |
264588_at |
AT2G17730
|
NIP2, NEP-interacting protein 2 |
0.408 |
-0.029 |
| 52 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-0.331 |
0.054 |
| 53 |
261684_at |
AT1G47400
|
unknown |
-0.283 |
-0.201 |
| 54 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.270 |
-0.078 |
| 55 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.230 |
-0.098 |
| 56 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.214 |
0.012 |
| 57 |
248558_at |
AT5G49990
|
[Xanthine/uracil permease family protein] |
-0.195 |
-0.087 |
| 58 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.193 |
0.028 |
| 59 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.187 |
0.072 |
| 60 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.162 |
-0.050 |
| 61 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
-0.150 |
-0.289 |
| 62 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.148 |
-0.128 |
| 63 |
263104_at |
AT2G05120
|
[Nucleoporin, Nup133/Nup155-like] |
-0.144 |
0.030 |
| 64 |
245761_at |
AT1G66890
|
unknown |
-0.142 |
0.044 |
| 65 |
265912_at |
AT2G25570
|
[binding] |
-0.136 |
-0.086 |
| 66 |
259993_at |
AT1G67960
|
POD1, POLLEN DEFECTIVE IN GUIDANCE 1 |
-0.136 |
-0.044 |
| 67 |
249948_at |
AT5G19210
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.136 |
0.235 |
| 68 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.127 |
-0.152 |
| 69 |
251929_at |
AT3G53920
|
SIG3, SIGMA FACTOR 3, SIGC, RNApolymerase sigma-subunit C |
-0.121 |
-0.196 |
| 70 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
-0.120 |
0.023 |
| 71 |
261062_at |
AT1G07530
|
ATGRAS2, ARABIDOPSIS THALIANA GRAS (GAI, RGA, SCR) 2, GRAS2, GRAS (GAI, RGA, SCR) 2, SCL14, SCARECROW-like 14 |
-0.116 |
-0.019 |
| 72 |
249231_at |
AT5G42030
|
ABIL4, ABL interactor-like protein 4 |
-0.116 |
0.285 |
| 73 |
258673_at |
AT3G08620
|
[RNA-binding KH domain-containing protein] |
-0.116 |
-0.030 |
| 74 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
-0.113 |
-0.029 |
| 75 |
262067_at |
AT1G80060
|
[Ubiquitin-like superfamily protein] |
-0.113 |
-0.097 |
| 76 |
261652_at |
AT1G01860
|
PFC1, PALEFACE 1 |
-0.112 |
0.208 |
| 77 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
-0.110 |
0.037 |
| 78 |
253665_at |
AT4G30230
|
unknown |
-0.109 |
0.039 |
| 79 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
-0.105 |
0.042 |
| 80 |
248319_at |
AT5G52710
|
[Copper transport protein family] |
-0.104 |
0.042 |
| 81 |
253157_at |
AT4G35740
|
ATRECQ3, A. THALIANA RECQ HELICASE 3, RecQl3 |
-0.103 |
-0.119 |
| 82 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.101 |
-0.057 |
| 83 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.100 |
-0.031 |
| 84 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
-0.095 |
-0.104 |
| 85 |
255522_at |
AT4G02260
|
AT-RSH1, RELA-SPOT HOMOLOG 1, ATRSH1, RELA/SPOT HOMOLOG 1, RSH1, RELA/SPOT homolog 1 |
-0.094 |
-0.065 |
| 86 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.090 |
-0.082 |
| 87 |
264320_at |
AT1G04090
|
unknown |
-0.089 |
-0.000 |
| 88 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.089 |
0.101 |
| 89 |
246180_at |
AT5G20840
|
[Phosphoinositide phosphatase family protein] |
-0.088 |
0.026 |
| 90 |
248504_at |
AT5G50480
|
NF-YC6, nuclear factor Y, subunit C6 |
-0.087 |
-0.046 |
| 91 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
-0.087 |
0.065 |
| 92 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
-0.087 |
0.020 |
| 93 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.086 |
0.157 |
| 94 |
266126_at |
AT2G45040
|
[Matrixin family protein] |
-0.085 |
-0.196 |
| 95 |
261945_at |
AT1G64530
|
NLP6, NIN-like protein 6 |
-0.083 |
-0.011 |
| 96 |
263257_at |
AT1G10520
|
AtPol{lambda}, Pol{lambda}, DNA polymerase {lambda} |
-0.080 |
-0.000 |
| 97 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
-0.080 |
-0.077 |
| 98 |
262076_at |
AT1G59580
|
ATMPK2, mitogen-activated protein kinase homolog 2, MPK2, mitogen-activated protein kinase homolog 2 |
-0.078 |
0.057 |
| 99 |
253892_at |
AT4G27620
|
unknown |
-0.077 |
0.044 |
| 100 |
248003_at |
AT5G56220
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.075 |
-0.141 |
| 101 |
256410_at |
AT1G66630
|
[Protein with RING/U-box and TRAF-like domains] |
-0.071 |
0.012 |