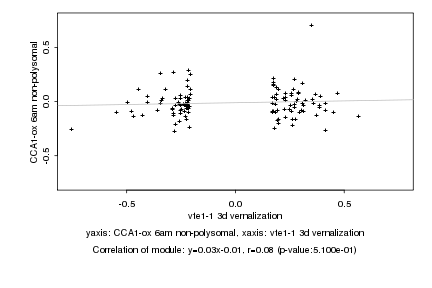
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
vte1-1 3d vernalization |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.564 |
-0.135 |
| 2 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.469 |
0.078 |
| 3 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.448 |
-0.098 |
| 4 |
256176_at |
AT1G51640
|
ATEXO70G2, exocyst subunit exo70 family protein G2, EXO70G2, exocyst subunit exo70 family protein G2 |
0.414 |
-0.076 |
| 5 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.413 |
-0.014 |
| 6 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.413 |
-0.267 |
| 7 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.390 |
0.047 |
| 8 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
0.386 |
-0.048 |
| 9 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.386 |
-0.029 |
| 10 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
0.372 |
-0.120 |
| 11 |
246125_at |
AT5G19875
|
unknown |
0.368 |
0.068 |
| 12 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
0.357 |
-0.011 |
| 13 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
0.351 |
0.026 |
| 14 |
264643_at |
AT1G08990
|
GUX5, Glucuronic Acid Substitution of Xylan 5, PGSIP5, plant glycogenin-like starch initiation protein 5 |
0.346 |
0.711 |
| 15 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
0.318 |
0.017 |
| 16 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
0.310 |
-0.087 |
| 17 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.310 |
-0.032 |
| 18 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.306 |
-0.018 |
| 19 |
249868_at |
AT5G23030
|
TET12, tetraspanin12 |
0.304 |
0.170 |
| 20 |
259735_at |
AT1G64405
|
unknown |
0.301 |
-0.074 |
| 21 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.292 |
-0.097 |
| 22 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.289 |
0.078 |
| 23 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.288 |
0.084 |
| 24 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.282 |
0.025 |
| 25 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
0.280 |
0.002 |
| 26 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
0.274 |
-0.158 |
| 27 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
0.269 |
-0.054 |
| 28 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.268 |
-0.025 |
| 29 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.267 |
0.209 |
| 30 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
0.264 |
0.119 |
| 31 |
260423_at |
AT1G72470
|
ATEXO70D1, exocyst subunit exo70 family protein D1, EXO70D1, exocyst subunit exo70 family protein D1 |
0.261 |
-0.219 |
| 32 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.259 |
-0.160 |
| 33 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
0.255 |
-0.092 |
| 34 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
0.255 |
0.081 |
| 35 |
250744_at |
AT5G05840
|
unknown |
0.254 |
0.056 |
| 36 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.249 |
-0.033 |
| 37 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
0.248 |
-0.069 |
| 38 |
254281_at |
AT4G22840
|
[Sodium Bile acid symporter family] |
0.229 |
0.079 |
| 39 |
251299_at |
AT3G61950
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.229 |
0.043 |
| 40 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.227 |
0.018 |
| 41 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
0.226 |
-0.142 |
| 42 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.223 |
-0.070 |
| 43 |
266968_at |
AT2G39360
|
[Protein kinase superfamily protein] |
0.220 |
0.036 |
| 44 |
264481_at |
AT1G77200
|
[Integrase-type DNA-binding superfamily protein] |
0.196 |
-0.161 |
| 45 |
251600_at |
AT3G57710
|
[Protein kinase superfamily protein] |
0.196 |
0.117 |
| 46 |
261684_at |
AT1G47400
|
unknown |
0.194 |
-0.201 |
| 47 |
251521_at |
AT3G59420
|
ACR4, crinkly4, CR4, crinkly4 |
0.192 |
-0.077 |
| 48 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
0.191 |
-0.167 |
| 49 |
263629_at |
AT2G04850
|
[Auxin-responsive family protein] |
0.188 |
0.138 |
| 50 |
249619_at |
AT5G37500
|
GORK, gated outwardly-rectifying K+ channel |
0.186 |
0.027 |
| 51 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.185 |
0.073 |
| 52 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
0.184 |
-0.100 |
| 53 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
0.181 |
-0.019 |
| 54 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
0.179 |
0.046 |
| 55 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.177 |
-0.240 |
| 56 |
247354_at |
AT5G63590
|
ATFLS3, FLS3, flavonol synthase 3 |
0.173 |
0.160 |
| 57 |
250998_at |
AT5G02620
|
ANK1, ankyrin-like1, ATANK1 |
0.172 |
0.152 |
| 58 |
247877_at |
AT5G57740
|
XBAT32, XB3 ortholog 2 in Arabidopsis thaliana |
0.171 |
-0.087 |
| 59 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.171 |
0.180 |
| 60 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
0.171 |
0.219 |
| 61 |
259606_at |
AT1G27920
|
MAP65-8, microtubule-associated protein 65-8 |
0.167 |
-0.095 |
| 62 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.167 |
0.038 |
| 63 |
255647_at |
AT4G00900
|
ATECA2, ARABIDOPSIS THALIANA ER-TYPE CA2+-ATPASE 2, ECA2, ER-type Ca2+-ATPase 2 |
0.167 |
-0.088 |
| 64 |
257124_at |
AT3G20040
|
ATHXK4, HKL2, HEXOKINASE-LIKE 2 |
0.167 |
-0.012 |
| 65 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.757 |
-0.255 |
| 66 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.547 |
-0.099 |
| 67 |
253394_at |
AT4G32770
|
ATSDX1, SUCROSE EXPORT DEFECTIVE 1, VTE1, VITAMIN E DEFICIENT 1 |
-0.500 |
-0.005 |
| 68 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-0.479 |
-0.091 |
| 69 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.469 |
-0.136 |
| 70 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.449 |
0.111 |
| 71 |
252539_at |
AT3G45730
|
unknown |
-0.428 |
-0.125 |
| 72 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.408 |
-0.005 |
| 73 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.406 |
0.052 |
| 74 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
-0.361 |
-0.074 |
| 75 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
-0.349 |
0.263 |
| 76 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.347 |
-0.015 |
| 77 |
250223_at |
AT5G14070
|
ROXY2 |
-0.344 |
0.017 |
| 78 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.336 |
0.028 |
| 79 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.323 |
0.114 |
| 80 |
248225_at |
AT5G53740
|
unknown |
-0.293 |
-0.069 |
| 81 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.290 |
-0.059 |
| 82 |
264379_at |
AT2G25200
|
unknown |
-0.287 |
0.277 |
| 83 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
-0.286 |
-0.107 |
| 84 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
-0.286 |
-0.123 |
| 85 |
262211_at |
AT1G74930
|
ORA47 |
-0.281 |
-0.276 |
| 86 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.279 |
0.035 |
| 87 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.279 |
-0.212 |
| 88 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
-0.276 |
-0.034 |
| 89 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.266 |
-0.002 |
| 90 |
260991_at |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
-0.261 |
-0.025 |
| 91 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.259 |
-0.177 |
| 92 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
-0.257 |
-0.106 |
| 93 |
261941_at |
AT1G22490
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.255 |
-0.075 |
| 94 |
254571_at |
AT4G19370
|
unknown |
-0.255 |
-0.033 |
| 95 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.253 |
0.059 |
| 96 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.253 |
0.032 |
| 97 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.252 |
-0.065 |
| 98 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
-0.243 |
-0.025 |
| 99 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.239 |
-0.024 |
| 100 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.238 |
-0.091 |
| 101 |
249017_at |
AT5G44760
|
[C2 domain-containing protein] |
-0.235 |
-0.028 |
| 102 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.233 |
-0.013 |
| 103 |
257398_at |
AT2G01990
|
unknown |
-0.233 |
-0.136 |
| 104 |
249791_at |
AT5G23810
|
AAP7, amino acid permease 7 |
-0.233 |
-0.019 |
| 105 |
245224_at |
AT3G29796
|
unknown |
-0.232 |
0.046 |
| 106 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.231 |
-0.024 |
| 107 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
-0.228 |
-0.160 |
| 108 |
255599_at |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
-0.227 |
-0.063 |
| 109 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.225 |
-0.038 |
| 110 |
259015_at |
AT3G07350
|
unknown |
-0.224 |
-0.021 |
| 111 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
-0.223 |
0.144 |
| 112 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.223 |
-0.001 |
| 113 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
-0.223 |
0.198 |
| 114 |
261213_at |
AT1G12970
|
PIRL3, plant intracellular ras group-related LRR 3 |
-0.222 |
0.014 |
| 115 |
246755_at |
AT5G27920
|
[F-box family protein] |
-0.221 |
-0.059 |
| 116 |
263475_at |
AT2G31945
|
unknown |
-0.221 |
0.041 |
| 117 |
246941_at |
AT5G25415
|
unknown |
-0.219 |
-0.100 |
| 118 |
261736_at |
AT1G47810
|
[F-box and associated interaction domains-containing protein] |
-0.219 |
0.287 |
| 119 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
-0.219 |
0.015 |
| 120 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.216 |
-0.038 |
| 121 |
257776_at |
AT3G29190
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.216 |
-0.063 |
| 122 |
261028_at |
AT1G26620
|
unknown |
-0.213 |
-0.041 |
| 123 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.212 |
-0.236 |
| 124 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
-0.212 |
0.036 |
| 125 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.211 |
0.253 |
| 126 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.208 |
0.067 |
| 127 |
254712_at |
AT4G18080
|
unknown |
-0.208 |
0.113 |