|
probeID |
AGICode |
Annotation |
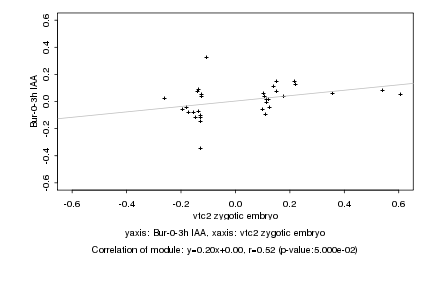
Log2 signal ratio
vtc2 zygotic embryo |
Log2 signal ratio
Bur-0-3h IAA |
| 1 |
252957_at |
AT4G38680
|
ATCSP2, ARABIDOPSIS THALIANA COLD SHOCK PROTEIN 2, CSDP2, COLD SHOCK DOMAIN PROTEIN 2, CSP2, COLD SHOCK PROTEIN 2, GRP2, glycine rich protein 2 |
0.605 |
0.052 |
| 2 |
258501_at |
AT3G06780
|
[glycine-rich protein] |
0.537 |
0.083 |
| 3 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.353 |
0.065 |
| 4 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.217 |
0.130 |
| 5 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.215 |
0.149 |
| 6 |
252197_at |
AT3G50230
|
[Leucine-rich repeat protein kinase family protein] |
0.176 |
0.041 |
| 7 |
261413_at |
AT1G07630
|
PLL5, pol-like 5 |
0.150 |
0.081 |
| 8 |
246796_at |
AT5G26770
|
unknown |
0.148 |
0.152 |
| 9 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
0.138 |
0.114 |
| 10 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.125 |
-0.042 |
| 11 |
247263_at |
AT5G64470
|
TBL12, TRICHOME BIREFRINGENCE-LIKE 12 |
0.120 |
0.022 |
| 12 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
0.114 |
-0.004 |
| 13 |
266117_at |
AT2G02170
|
[Remorin family protein] |
0.113 |
0.020 |
| 14 |
263528_at |
AT2G24800
|
[Peroxidase superfamily protein] |
0.110 |
-0.093 |
| 15 |
256292_at |
AT1G69430
|
unknown |
0.104 |
0.041 |
| 16 |
263887_at |
AT2G36980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.100 |
0.060 |
| 17 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
0.099 |
-0.055 |
| 18 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
-0.261 |
0.024 |
| 19 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
-0.195 |
-0.052 |
| 20 |
261181_at |
AT1G34580
|
[Major facilitator superfamily protein] |
-0.182 |
-0.043 |
| 21 |
247611_at |
AT5G60710
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.173 |
-0.077 |
| 22 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
-0.156 |
-0.078 |
| 23 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.149 |
-0.113 |
| 24 |
257480_at |
AT1G15640
|
unknown |
-0.140 |
0.080 |
| 25 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.137 |
0.092 |
| 26 |
247627_at |
AT5G60360
|
AALP, aleurain-like protease, ALP, aleurain-like protease, SAG2, SENESCENCE ASSOCIATED GENE2 |
-0.136 |
-0.067 |
| 27 |
252720_at |
AT3G43970
|
unknown |
-0.132 |
-0.097 |
| 28 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.132 |
-0.141 |
| 29 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.132 |
-0.115 |
| 30 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.131 |
-0.341 |
| 31 |
254100_at |
AT4G25020
|
[D111/G-patch domain-containing protein] |
-0.128 |
0.052 |
| 32 |
249274_at |
AT5G41860
|
unknown |
-0.128 |
0.040 |
| 33 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
-0.110 |
0.330 |