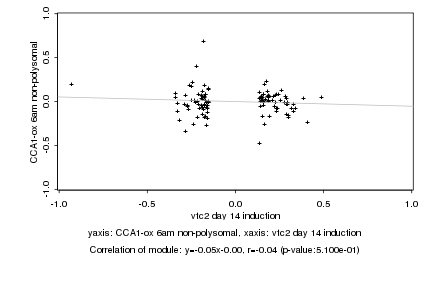
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
vtc2 day 14 induction |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
267431_at |
AT2G34870
|
MEE26, maternal effect embryo arrest 26 |
0.487 |
0.046 |
| 2 |
253241_at |
AT4G34520
|
FAE1, FATTY ACID ELONGATION1, KCS18, 3-ketoacyl-CoA synthase 18 |
0.408 |
-0.234 |
| 3 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
0.385 |
0.035 |
| 4 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.338 |
-0.070 |
| 5 |
248431_at |
AT5G51470
|
[Auxin-responsive GH3 family protein] |
0.328 |
-0.032 |
| 6 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
0.324 |
-0.105 |
| 7 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.317 |
-0.073 |
| 8 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.300 |
-0.178 |
| 9 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
0.299 |
-0.156 |
| 10 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.293 |
-0.005 |
| 11 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
0.291 |
-0.031 |
| 12 |
254264_at |
AT4G23510
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.288 |
0.045 |
| 13 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.288 |
-0.026 |
| 14 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.287 |
-0.143 |
| 15 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.279 |
0.068 |
| 16 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
0.274 |
-0.004 |
| 17 |
253150_at |
AT4G35660
|
unknown |
0.259 |
0.131 |
| 18 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
0.251 |
0.016 |
| 19 |
249206_at |
AT5G42630
|
ATS, ABERRANT TESTA SHAPE, KAN4, KANADI 4 |
0.239 |
0.091 |
| 20 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
0.236 |
-0.072 |
| 21 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.231 |
0.082 |
| 22 |
258760_at |
AT3G10780
|
[emp24/gp25L/p24 family/GOLD family protein] |
0.228 |
-0.078 |
| 23 |
248647_at |
AT5G49190
|
ATSUS2, SSA, SUCROSE SYNTHASE FROM ARABIDOPSIS, SUS2, sucrose synthase 2 |
0.228 |
-0.105 |
| 24 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.223 |
0.071 |
| 25 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.222 |
-0.003 |
| 26 |
263042_at |
AT1G23340
|
unknown |
0.221 |
-0.049 |
| 27 |
264721_at |
AT1G23000
|
[Heavy metal transport/detoxification superfamily protein ] |
0.212 |
0.061 |
| 28 |
248754_at |
AT5G47670
|
L1L, LEC1-LIKE, NF-YB6, nuclear factor Y, subunit B6 |
0.209 |
0.022 |
| 29 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.193 |
0.058 |
| 30 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.192 |
-0.160 |
| 31 |
260542_at |
AT2G43560
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.187 |
0.079 |
| 32 |
260639_at |
AT1G53180
|
unknown |
0.185 |
0.019 |
| 33 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
0.185 |
0.007 |
| 34 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
0.184 |
0.046 |
| 35 |
245075_at |
AT2G23180
|
CYP96A1, cytochrome P450, family 96, subfamily A, polypeptide 1 |
0.182 |
0.002 |
| 36 |
257229_at |
AT3G16490
|
IQD26, IQ-domain 26 |
0.180 |
0.120 |
| 37 |
262278_at |
AT1G68640
|
PAN, PERIANTHIA, TGA8, TGACG sequence-specific binding protein 8 |
0.180 |
0.067 |
| 38 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
0.176 |
0.231 |
| 39 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
0.169 |
0.029 |
| 40 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
0.164 |
-0.253 |
| 41 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.162 |
0.194 |
| 42 |
264124_at |
AT1G79360
|
ATOCT2, organic cation/carnitine transporter 2, OCT2, ORGANIC CATION TRANSPORTER 2, OCT2, organic cation/carnitine transporter 2 |
0.159 |
0.002 |
| 43 |
263352_at |
AT2G22080
|
unknown |
0.158 |
0.081 |
| 44 |
245161_at |
AT2G33070
|
ATNSP2, NITRILE-SPECIFIER PROTEIN 2, NSP2, nitrile specifier protein 2 |
0.157 |
-0.007 |
| 45 |
257285_at |
AT3G29760
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.154 |
-0.038 |
| 46 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.154 |
0.083 |
| 47 |
249716_at |
AT5G35770
|
SAP, STERILE APETALA |
0.153 |
0.018 |
| 48 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.152 |
0.053 |
| 49 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.151 |
0.023 |
| 50 |
264517_at |
AT1G10120
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.151 |
-0.168 |
| 51 |
251081_at |
AT5G02070
|
[Protein kinase family protein] |
0.144 |
0.039 |
| 52 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.144 |
0.016 |
| 53 |
246125_at |
AT5G19875
|
unknown |
0.143 |
0.068 |
| 54 |
256212_at |
AT1G50970
|
[Membrane trafficking VPS53 family protein] |
0.142 |
-0.046 |
| 55 |
261260_at |
AT1G26680
|
[transcriptional factor B3 family protein] |
0.141 |
0.053 |
| 56 |
252248_at |
AT3G49750
|
AtRLP44, receptor like protein 44, RLP44, receptor like protein 44 |
0.137 |
0.009 |
| 57 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
0.134 |
0.037 |
| 58 |
257698_at |
AT3G12730
|
[Homeodomain-like superfamily protein] |
0.133 |
0.105 |
| 59 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
0.132 |
-0.470 |
| 60 |
253922_at |
AT4G26850
|
VTC2, vitamin c defective 2 |
-0.934 |
0.195 |
| 61 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.344 |
0.096 |
| 62 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.343 |
0.050 |
| 63 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.331 |
-0.022 |
| 64 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.329 |
-0.109 |
| 65 |
245233_at |
AT4G25580
|
[CAP160 protein] |
-0.319 |
-0.216 |
| 66 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.294 |
-0.033 |
| 67 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.288 |
-0.333 |
| 68 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.287 |
0.072 |
| 69 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.276 |
-0.050 |
| 70 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.273 |
-0.045 |
| 71 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.267 |
-0.088 |
| 72 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
-0.262 |
0.192 |
| 73 |
253936_at |
AT4G26880
|
[Stigma-specific Stig1 family protein] |
-0.254 |
0.016 |
| 74 |
250826_at |
AT5G05220
|
unknown |
-0.252 |
0.177 |
| 75 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.248 |
0.228 |
| 76 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.239 |
-0.254 |
| 77 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.236 |
0.017 |
| 78 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
-0.231 |
-0.005 |
| 79 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
-0.223 |
0.409 |
| 80 |
263492_at |
AT2G42560
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.220 |
-0.181 |
| 81 |
256358_at |
AT1G66470
|
AtRHD6, RHD6, ROOT HAIR DEFECTIVE6 |
-0.216 |
0.001 |
| 82 |
266611_at |
AT2G14960
|
GH3.1 |
-0.215 |
0.083 |
| 83 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-0.212 |
-0.024 |
| 84 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.206 |
-0.069 |
| 85 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
-0.198 |
0.037 |
| 86 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
-0.196 |
-0.042 |
| 87 |
258596_at |
AT3G04510
|
LSH2, LIGHT SENSITIVE HYPOCOTYLS 2 |
-0.195 |
0.079 |
| 88 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
-0.191 |
-0.139 |
| 89 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
-0.191 |
-0.066 |
| 90 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.190 |
0.059 |
| 91 |
257059_at |
AT3G15280
|
unknown |
-0.190 |
-0.059 |
| 92 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.189 |
0.032 |
| 93 |
252914_at |
AT4G39130
|
[Dehydrin family protein] |
-0.187 |
0.125 |
| 94 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.186 |
0.690 |
| 95 |
249111_at |
AT5G43770
|
[proline-rich family protein] |
-0.185 |
-0.080 |
| 96 |
255596_at |
AT4G01720
|
AtWRKY47, WRKY47 |
-0.181 |
0.059 |
| 97 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
-0.181 |
-0.023 |
| 98 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.180 |
-0.044 |
| 99 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.177 |
0.191 |
| 100 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.176 |
0.046 |
| 101 |
247645_at |
AT5G60530
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.176 |
-0.179 |
| 102 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.175 |
0.014 |
| 103 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
-0.172 |
0.086 |
| 104 |
264480_at |
AT1G77300
|
ASHH2, ASH1 HOMOLOG 2, CCR1, CAROTENOID CHLOROPLAST REGULATORY1, EFS, EARLY FLOWERING IN SHORT DAYS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 |
-0.172 |
-0.169 |
| 105 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-0.170 |
0.083 |
| 106 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.167 |
-0.068 |
| 107 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.166 |
-0.001 |
| 108 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.165 |
-0.264 |
| 109 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.164 |
-0.193 |
| 110 |
254474_at |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
-0.163 |
-0.041 |
| 111 |
262049_at |
AT1G80180
|
unknown |
-0.161 |
-0.125 |
| 112 |
266274_at |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
-0.159 |
-0.069 |
| 113 |
250622_at |
AT5G07310
|
[Integrase-type DNA-binding superfamily protein] |
-0.157 |
-0.006 |
| 114 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.156 |
0.146 |
| 115 |
266170_at |
AT2G39050
|
ArathEULS3, EULS3, Euonymus lectin S3 |
-0.156 |
-0.006 |
| 116 |
263866_at |
AT2G36950
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.155 |
0.151 |
| 117 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
-0.153 |
0.007 |