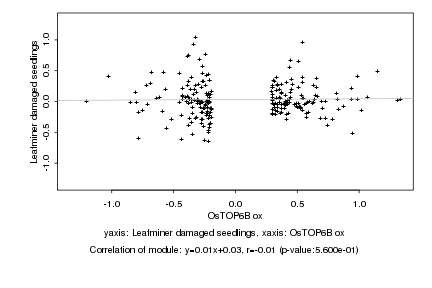
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
OsTOP6B ox |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
266393_at |
AT2G41260
|
ATM17, M17 |
1.331 |
0.037 |
| 2 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
1.309 |
0.021 |
| 3 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
1.148 |
0.490 |
| 4 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
1.062 |
0.077 |
| 5 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
1.015 |
-0.142 |
| 6 |
263023_at |
AT1G23960
|
unknown |
0.985 |
0.040 |
| 7 |
256021_at |
AT1G58270
|
ZW9 |
0.984 |
0.411 |
| 8 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.943 |
-0.503 |
| 9 |
265831_at |
AT2G14460
|
unknown |
0.934 |
0.043 |
| 10 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.932 |
0.212 |
| 11 |
252659_at |
AT3G44430
|
unknown |
0.872 |
-0.076 |
| 12 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.832 |
-0.116 |
| 13 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
0.824 |
0.048 |
| 14 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.814 |
0.136 |
| 15 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.782 |
-0.277 |
| 16 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.741 |
-0.383 |
| 17 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
0.727 |
0.004 |
| 18 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.725 |
-0.270 |
| 19 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.696 |
-0.099 |
| 20 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
0.692 |
0.011 |
| 21 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.686 |
-0.275 |
| 22 |
265297_at |
AT2G14080
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.662 |
0.087 |
| 23 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.652 |
0.382 |
| 24 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
0.652 |
0.231 |
| 25 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.641 |
0.098 |
| 26 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
0.634 |
0.034 |
| 27 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.628 |
-0.005 |
| 28 |
246888_at |
AT5G26270
|
unknown |
0.625 |
0.273 |
| 29 |
262010_at |
AT1G35612
|
[expressed protein] |
0.619 |
-0.027 |
| 30 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
0.598 |
0.005 |
| 31 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.585 |
-0.170 |
| 32 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.578 |
-0.003 |
| 33 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.574 |
-0.251 |
| 34 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.572 |
-0.187 |
| 35 |
248079_at |
AT5G55790
|
unknown |
0.571 |
-0.024 |
| 36 |
254724_at |
AT4G13630
|
unknown |
0.555 |
-0.046 |
| 37 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.550 |
0.053 |
| 38 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.538 |
0.967 |
| 39 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
0.537 |
0.322 |
| 40 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.537 |
0.065 |
| 41 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
0.536 |
0.390 |
| 42 |
251165_at |
AT3G63340
|
[Protein phosphatase 2C family protein] |
0.536 |
-0.139 |
| 43 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.528 |
0.108 |
| 44 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
0.521 |
-0.086 |
| 45 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.521 |
-0.093 |
| 46 |
249592_at |
AT5G37890
|
[Protein with RING/U-box and TRAF-like domains] |
0.520 |
-0.099 |
| 47 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
0.513 |
-0.079 |
| 48 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.508 |
0.243 |
| 49 |
265439_at |
AT2G21045
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.504 |
-0.028 |
| 50 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.502 |
0.654 |
| 51 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
0.497 |
-0.085 |
| 52 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.493 |
-0.069 |
| 53 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
0.493 |
-0.017 |
| 54 |
256132_at |
AT1G13610
|
[alpha/beta-Hydrolases superfamily protein] |
0.471 |
-0.034 |
| 55 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
0.460 |
0.282 |
| 56 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.453 |
0.369 |
| 57 |
248192_at |
AT5G54140
|
ILL3, IAA-leucine-resistant (ILR1)-like 3 |
0.448 |
0.196 |
| 58 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.441 |
0.678 |
| 59 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.441 |
0.159 |
| 60 |
264874_at |
AT1G24240
|
[Ribosomal protein L19 family protein] |
0.439 |
0.078 |
| 61 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.435 |
-0.012 |
| 62 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.434 |
0.558 |
| 63 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.432 |
0.039 |
| 64 |
258029_at |
AT3G27580
|
ATPK7, D6PKL3, D6 PROTEIN KINASE LIKE 3 |
0.426 |
-0.184 |
| 65 |
246699_at |
AT5G27990
|
[Pre-rRNA-processing protein TSR2, conserved region] |
0.419 |
0.022 |
| 66 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
0.418 |
0.023 |
| 67 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.413 |
-0.044 |
| 68 |
254455_at |
AT4G21140
|
unknown |
0.413 |
-0.010 |
| 69 |
245106_at |
AT2G41650
|
unknown |
0.411 |
-0.026 |
| 70 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.410 |
-0.287 |
| 71 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.409 |
0.318 |
| 72 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
0.405 |
-0.203 |
| 73 |
254476_at |
AT4G20410
|
GAMMA-SNAP, GAMMA-SOLUBLE NSF ATTACHMENT PROTEIN, GSNAP, gamma-soluble NSF attachment protein |
0.400 |
0.109 |
| 74 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
0.399 |
0.007 |
| 75 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.399 |
-0.015 |
| 76 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
0.399 |
-0.057 |
| 77 |
258045_at |
AT3G21280
|
UBP7, ubiquitin-specific protease 7 |
0.397 |
-0.022 |
| 78 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
0.395 |
-0.040 |
| 79 |
251600_at |
AT3G57710
|
[Protein kinase superfamily protein] |
0.391 |
-0.019 |
| 80 |
248940_at |
AT5G45400
|
ATRPA70C, RPA70C |
0.390 |
-0.108 |
| 81 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.372 |
-0.111 |
| 82 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.371 |
-0.173 |
| 83 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.368 |
0.291 |
| 84 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.368 |
-0.039 |
| 85 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
0.366 |
0.156 |
| 86 |
258107_at |
AT3G23560
|
ALF5, ABERRANT LATERAL ROOT FORMATION 5 |
0.365 |
0.132 |
| 87 |
263873_at |
AT2G21860
|
[violaxanthin de-epoxidase-related] |
0.362 |
-0.181 |
| 88 |
257322_at |
ATMG01180
|
ORF111B |
0.358 |
0.051 |
| 89 |
257423_at |
AT1G62010
|
[Mitochondrial transcription termination factor family protein] |
0.355 |
-0.001 |
| 90 |
252882_at |
AT4G39675
|
unknown |
0.354 |
0.080 |
| 91 |
252419_at |
AT3G47510
|
unknown |
0.353 |
0.198 |
| 92 |
252240_at |
AT3G50010
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.352 |
-0.013 |
| 93 |
248181_at |
AT5G54290
|
CcdA |
0.335 |
-0.090 |
| 94 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
0.334 |
0.274 |
| 95 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.334 |
-0.161 |
| 96 |
262873_at |
AT1G64700
|
unknown |
0.332 |
-0.041 |
| 97 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.332 |
0.286 |
| 98 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.331 |
0.063 |
| 99 |
257432_at |
AT2G21850
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.331 |
0.061 |
| 100 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.326 |
0.189 |
| 101 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.325 |
0.186 |
| 102 |
254200_at |
AT4G24110
|
unknown |
0.324 |
0.395 |
| 103 |
254402_at |
AT4G21310
|
unknown |
0.322 |
-0.131 |
| 104 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
0.316 |
-0.209 |
| 105 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
0.316 |
-0.018 |
| 106 |
246808_at |
AT5G27110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.313 |
-0.087 |
| 107 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.313 |
0.340 |
| 108 |
255858_at |
AT1G67030
|
ZFP6, zinc finger protein 6 |
0.311 |
-0.036 |
| 109 |
265341_at |
AT2G18360
|
[alpha/beta-Hydrolases superfamily protein] |
0.304 |
-0.048 |
| 110 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.301 |
0.353 |
| 111 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.297 |
0.272 |
| 112 |
259495_at |
AT1G15890
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.297 |
0.232 |
| 113 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
0.296 |
0.031 |
| 114 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.296 |
-0.189 |
| 115 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
0.296 |
0.130 |
| 116 |
258170_at |
AT3G21600
|
[Senescence/dehydration-associated protein-related] |
0.296 |
0.049 |
| 117 |
258027_at |
AT3G19515
|
unknown |
0.295 |
-0.197 |
| 118 |
245944_at |
AT5G19520
|
ATMSL9, MSL9, mechanosensitive channel of small conductance-like 9 |
0.295 |
-0.032 |
| 119 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.294 |
0.067 |
| 120 |
257410_at |
AT1G24340
|
EMB2421, EMBRYO DEFECTIVE 2421, EMB260, EMBRYO DEFECTIVE 260 |
0.293 |
-0.128 |
| 121 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.292 |
0.228 |
| 122 |
258646_at |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
0.292 |
-0.188 |
| 123 |
259952_at |
AT1G71400
|
AtRLP12, receptor like protein 12, RLP12, receptor like protein 12 |
0.290 |
0.164 |
| 124 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.206 |
0.002 |
| 125 |
252345_at |
AT3G48640
|
unknown |
-1.028 |
0.420 |
| 126 |
250135_at |
AT5G15360
|
unknown |
-0.850 |
-0.011 |
| 127 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.809 |
0.162 |
| 128 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.802 |
-0.013 |
| 129 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.789 |
-0.169 |
| 130 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.787 |
-0.584 |
| 131 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.752 |
-0.138 |
| 132 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.722 |
0.275 |
| 133 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-0.711 |
-0.040 |
| 134 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.694 |
0.300 |
| 135 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.679 |
0.487 |
| 136 |
254582_at |
AT4G19470
|
[Leucine-rich repeat (LRR) family protein] |
-0.641 |
0.063 |
| 137 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.617 |
0.072 |
| 138 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.590 |
-0.159 |
| 139 |
259385_at |
AT1G13470
|
unknown |
-0.588 |
0.479 |
| 140 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.572 |
0.197 |
| 141 |
263840_at |
AT2G36885
|
unknown |
-0.561 |
-0.426 |
| 142 |
259968_at |
AT1G76530
|
[Auxin efflux carrier family protein] |
-0.520 |
-0.281 |
| 143 |
248431_at |
AT5G51470
|
[Auxin-responsive GH3 family protein] |
-0.458 |
-0.013 |
| 144 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.455 |
0.456 |
| 145 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.441 |
-0.217 |
| 146 |
256096_at |
AT1G13650
|
unknown |
-0.437 |
-0.608 |
| 147 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.435 |
0.048 |
| 148 |
262126_at |
AT1G59620
|
CW9 |
-0.435 |
0.109 |
| 149 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.431 |
0.091 |
| 150 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
-0.428 |
0.213 |
| 151 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.408 |
0.072 |
| 152 |
246326_at |
AT1G16590
|
ATREV7, REV7 |
-0.399 |
0.003 |
| 153 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.395 |
0.263 |
| 154 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.393 |
0.089 |
| 155 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.389 |
0.732 |
| 156 |
246316_at |
AT3G56890
|
[F-box associated ubiquitination effector family protein] |
-0.386 |
-0.029 |
| 157 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.385 |
-0.386 |
| 158 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.384 |
0.081 |
| 159 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.382 |
0.333 |
| 160 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
-0.382 |
0.758 |
| 161 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.379 |
-0.268 |
| 162 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.376 |
0.048 |
| 163 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.369 |
0.210 |
| 164 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.362 |
0.042 |
| 165 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.360 |
-0.330 |
| 166 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.360 |
-0.007 |
| 167 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.357 |
0.398 |
| 168 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.354 |
-0.522 |
| 169 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
-0.352 |
-0.082 |
| 170 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.348 |
-0.192 |
| 171 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.347 |
0.931 |
| 172 |
256210_at |
AT1G50950
|
[INVOLVED IN: cell redox homeostasis] |
-0.347 |
0.122 |
| 173 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
-0.339 |
0.198 |
| 174 |
251030_at |
AT5G02130
|
NDP1 |
-0.333 |
0.023 |
| 175 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.327 |
0.265 |
| 176 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.324 |
-0.265 |
| 177 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.323 |
1.055 |
| 178 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.322 |
0.161 |
| 179 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.316 |
-0.245 |
| 180 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.311 |
-0.025 |
| 181 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.307 |
0.015 |
| 182 |
255142_at |
AT4G08390
|
SAPX, stromal ascorbate peroxidase |
-0.307 |
0.036 |
| 183 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
-0.306 |
0.025 |
| 184 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.301 |
0.288 |
| 185 |
251046_at |
AT5G02370
|
[ATP binding microtubule motor family protein] |
-0.295 |
-0.030 |
| 186 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.293 |
0.689 |
| 187 |
266412_at |
AT2G38720
|
MAP65-5, microtubule-associated protein 65-5 |
-0.289 |
-0.003 |
| 188 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.285 |
-0.028 |
| 189 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
-0.285 |
-0.097 |
| 190 |
250151_at |
AT5G14570
|
ATNRT2.7, high affinity nitrate transporter 2.7, NRT2.7, high affinity nitrate transporter 2.7 |
-0.280 |
-0.013 |
| 191 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.277 |
-0.284 |
| 192 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
-0.277 |
0.230 |
| 193 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.276 |
0.005 |
| 194 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.275 |
-0.348 |
| 195 |
257899_at |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
-0.275 |
-0.168 |
| 196 |
264738_at |
AT1G62250
|
unknown |
-0.274 |
-0.289 |
| 197 |
257925_at |
AT3G23170
|
unknown |
-0.273 |
0.469 |
| 198 |
263831_at |
AT2G40300
|
ATFER4, ferritin 4, FER4, ferritin 4 |
-0.270 |
-0.176 |
| 199 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.269 |
0.339 |
| 200 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
-0.269 |
-0.015 |
| 201 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.267 |
0.575 |
| 202 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.264 |
-0.112 |
| 203 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.264 |
-0.107 |
| 204 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.261 |
-0.404 |
| 205 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
-0.261 |
0.328 |
| 206 |
263674_at |
AT2G04790
|
unknown |
-0.253 |
-0.631 |
| 207 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
-0.252 |
-0.106 |
| 208 |
260005_at |
AT1G67920
|
unknown |
-0.248 |
0.768 |
| 209 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
-0.246 |
-0.048 |
| 210 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
-0.245 |
0.271 |
| 211 |
247319_at |
AT5G64050
|
ATERS, ERS, glutamate tRNA synthetase, OVA3, OVULE ABORTION 3 |
-0.244 |
-0.064 |
| 212 |
254290_at |
AT4G23000
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.242 |
-0.223 |
| 213 |
251243_at |
AT3G61870
|
unknown |
-0.241 |
-0.184 |
| 214 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.239 |
0.013 |
| 215 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.238 |
0.437 |
| 216 |
265539_at |
AT2G15830
|
unknown |
-0.238 |
0.140 |
| 217 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.232 |
0.012 |
| 218 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.232 |
0.025 |
| 219 |
247832_at |
AT5G58550
|
EOL2, ETO1-like 2 |
-0.231 |
-0.188 |
| 220 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.231 |
-0.090 |
| 221 |
261500_at |
AT1G28400
|
unknown |
-0.226 |
0.100 |
| 222 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.226 |
-0.147 |
| 223 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.224 |
0.141 |
| 224 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.224 |
-0.265 |
| 225 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
-0.223 |
-0.020 |
| 226 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.222 |
-0.455 |
| 227 |
246001_at |
AT5G20790
|
unknown |
-0.222 |
-0.646 |
| 228 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.220 |
-0.488 |
| 229 |
251191_at |
AT3G62590
|
[alpha/beta-Hydrolases superfamily protein] |
-0.218 |
0.454 |
| 230 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
-0.217 |
-0.211 |
| 231 |
254850_at |
AT4G12000
|
[SNARE associated Golgi protein family] |
-0.217 |
0.073 |
| 232 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
-0.213 |
0.347 |
| 233 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.213 |
0.350 |
| 234 |
253397_at |
AT4G32710
|
PERK14, proline-rich extensin-like receptor kinase 14 |
-0.213 |
-0.364 |
| 235 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.212 |
-0.418 |
| 236 |
260801_at |
AT1G78430
|
RIP4, ROP interactive partner 4 |
-0.211 |
-0.003 |
| 237 |
258802_at |
AT3G04650
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.210 |
0.011 |
| 238 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.209 |
-0.106 |
| 239 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.208 |
-0.352 |
| 240 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
-0.206 |
-0.344 |
| 241 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
-0.205 |
0.116 |
| 242 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.203 |
-0.175 |
| 243 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
-0.201 |
-0.256 |
| 244 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.198 |
-0.103 |
| 245 |
264954_at |
AT1G77060
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.198 |
0.167 |
| 246 |
246394_at |
AT1G58160
|
JAX1, JACALIN-TYPE LECTIN REQUIRED FOR POTEXVIRUS RESISTANCE 1 |
-0.197 |
-0.114 |