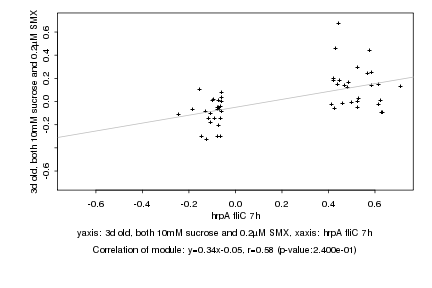
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
hrpA fliC 7h |
Log2 signal ratio
3d old, both 10mM sucrose and 0.2µM SMX |
| 1 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.708 |
0.133 |
| 2 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.631 |
-0.092 |
| 3 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.626 |
-0.089 |
| 4 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.622 |
0.017 |
| 5 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.615 |
-0.019 |
| 6 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.611 |
0.153 |
| 7 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.584 |
0.140 |
| 8 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
0.584 |
0.252 |
| 9 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.575 |
0.449 |
| 10 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.564 |
0.248 |
| 11 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
0.528 |
0.034 |
| 12 |
254249_at |
AT4G23280
|
CRK20, cysteine-rich RLK (RECEPTOR-like protein kinase) 20 |
0.522 |
0.001 |
| 13 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.522 |
-0.047 |
| 14 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.522 |
0.297 |
| 15 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.495 |
-0.001 |
| 16 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
0.484 |
0.168 |
| 17 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
0.479 |
0.123 |
| 18 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.468 |
0.140 |
| 19 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.459 |
-0.017 |
| 20 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.446 |
0.185 |
| 21 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.441 |
0.682 |
| 22 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.435 |
0.147 |
| 23 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.426 |
0.466 |
| 24 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.425 |
-0.052 |
| 25 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.421 |
0.183 |
| 26 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.419 |
0.205 |
| 27 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.412 |
-0.024 |
| 28 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
-0.246 |
-0.112 |
| 29 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.188 |
-0.061 |
| 30 |
263738_at |
AT1G60060
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
-0.155 |
0.106 |
| 31 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.149 |
-0.299 |
| 32 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.131 |
-0.078 |
| 33 |
267462_at |
AT2G33735
|
[Chaperone DnaJ-domain superfamily protein] |
-0.127 |
-0.320 |
| 34 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.120 |
-0.145 |
| 35 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.108 |
-0.102 |
| 36 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.108 |
-0.176 |
| 37 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
-0.101 |
0.012 |
| 38 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.097 |
0.019 |
| 39 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.093 |
-0.141 |
| 40 |
256111_at |
AT1G16820
|
[vacuolar ATP synthase catalytic subunit-related / V-ATPase-related / vacuolar proton pump-related] |
-0.079 |
-0.044 |
| 41 |
253145_at |
AT4G35560
|
DAW1, DUO1-activated WD40 1 |
-0.079 |
-0.296 |
| 42 |
257660_at |
AT3G13360
|
WIP3, WPP domain interacting protein 3 |
-0.078 |
-0.063 |
| 43 |
251387_at |
AT3G60810
|
unknown |
-0.076 |
-0.202 |
| 44 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.075 |
0.013 |
| 45 |
266120_at |
AT2G02070
|
AtIDD5, indeterminate(ID)-domain 5, IDD5, indeterminate(ID)-domain 5 |
-0.075 |
-0.051 |
| 46 |
267498_at |
AT2G45720
|
[ARM repeat superfamily protein] |
-0.069 |
-0.061 |
| 47 |
261417_at |
AT1G07700
|
[Thioredoxin superfamily protein] |
-0.068 |
-0.295 |
| 48 |
249568_at |
AT5G38040
|
[UDP-Glycosyltransferase superfamily protein] |
-0.066 |
-0.142 |
| 49 |
261616_at |
AT1G33060
|
ANAC014, NAC 014, NAC014, NAC 014 |
-0.066 |
-0.039 |
| 50 |
253225_at |
AT4G35020
|
ARAC3, RAC-like 3, ATROP6, RAC3, RAC-like 3, RHO1PS, ROP6, RHO-RELATED PROTEIN FROM PLANTS 6 |
-0.064 |
0.039 |
| 51 |
249565_at |
AT5G38440
|
[Plant self-incompatibility protein S1 family] |
-0.062 |
0.005 |
| 52 |
248835_at |
AT5G47250
|
[LRR and NB-ARC domains-containing disease resistance protein] |
-0.061 |
0.082 |
| 53 |
247742_at |
AT5G58980
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.061 |
-0.079 |