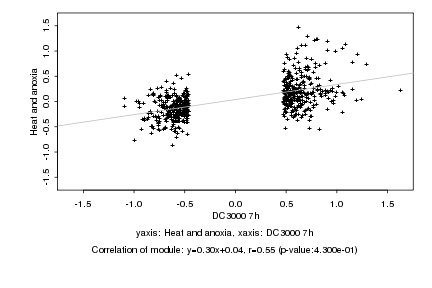
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
DC3000 7h |
Log2 signal ratio
Heat and anoxia |
| 1 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
1.624 |
0.220 |
| 2 |
266486_at |
AT2G47950
|
unknown |
1.284 |
0.752 |
| 3 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.237 |
0.046 |
| 4 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
1.198 |
0.944 |
| 5 |
247264_at |
AT5G64530
|
ANAC104, Arabidopsis NAC domain containing protein 104, XND1, xylem NAC domain 1 |
1.186 |
0.040 |
| 6 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
1.154 |
0.243 |
| 7 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
1.151 |
0.791 |
| 8 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.079 |
1.137 |
| 9 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
1.067 |
0.132 |
| 10 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
1.059 |
0.162 |
| 11 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
1.053 |
1.054 |
| 12 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.048 |
0.180 |
| 13 |
266456_at |
AT2G22770
|
NAI1 |
1.046 |
-0.204 |
| 14 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
1.015 |
0.315 |
| 15 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.989 |
0.186 |
| 16 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.982 |
0.995 |
| 17 |
267206_at |
AT2G30830
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.978 |
0.106 |
| 18 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
0.967 |
0.035 |
| 19 |
257418_at |
AT1G30850
|
RSH4, root hair specific 4 |
0.965 |
-0.031 |
| 20 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
0.957 |
0.207 |
| 21 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.954 |
0.258 |
| 22 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.941 |
0.010 |
| 23 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
0.937 |
0.237 |
| 24 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.928 |
0.436 |
| 25 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.914 |
0.188 |
| 26 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
0.910 |
0.077 |
| 27 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.907 |
-0.140 |
| 28 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.907 |
0.210 |
| 29 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.906 |
1.020 |
| 30 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.899 |
1.205 |
| 31 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.884 |
0.224 |
| 32 |
266455_at |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.881 |
0.124 |
| 33 |
260278_at |
AT1G80590
|
ATWRKY66, WRKY66, WRKY DNA-binding protein 66 |
0.881 |
0.175 |
| 34 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
0.881 |
0.761 |
| 35 |
258377_at |
AT3G17690
|
ATCNGC19, CYCLIC NUCLEOTIDE-GATED CHANNEL 19, CNGC19, cyclic nucleotide gated channel 19 |
0.851 |
0.204 |
| 36 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.844 |
0.110 |
| 37 |
256364_at |
AT1G66550
|
ATWRKY67, WRKY67, WRKY DNA-binding protein 67 |
0.836 |
0.257 |
| 38 |
265632_at |
AT2G14290
|
unknown |
0.835 |
0.152 |
| 39 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.833 |
0.160 |
| 40 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.825 |
-0.546 |
| 41 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.825 |
0.332 |
| 42 |
245209_at |
AT5G12340
|
unknown |
0.823 |
0.730 |
| 43 |
254200_at |
AT4G24110
|
unknown |
0.814 |
0.459 |
| 44 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.809 |
0.302 |
| 45 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.807 |
1.247 |
| 46 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.804 |
0.257 |
| 47 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.800 |
0.236 |
| 48 |
248090_at |
AT5G55090
|
MAPKKK15, mitogen-activated protein kinase kinase kinase 15 |
0.799 |
0.457 |
| 49 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.798 |
-0.055 |
| 50 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.794 |
1.242 |
| 51 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.787 |
0.762 |
| 52 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.781 |
0.677 |
| 53 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.775 |
0.029 |
| 54 |
247041_at |
AT5G67180
|
TOE3, target of early activation tagged (EAT) 3 |
0.775 |
0.110 |
| 55 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.775 |
1.212 |
| 56 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.768 |
-0.105 |
| 57 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.768 |
0.588 |
| 58 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
0.764 |
-0.066 |
| 59 |
265479_at |
AT2G15760
|
unknown |
0.762 |
0.188 |
| 60 |
247913_at |
AT5G57510
|
unknown |
0.762 |
0.258 |
| 61 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.751 |
0.188 |
| 62 |
257450_at |
AT1G10530
|
unknown |
0.750 |
0.153 |
| 63 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
0.749 |
0.848 |
| 64 |
259043_at |
AT3G03440
|
[ARM repeat superfamily protein] |
0.747 |
0.360 |
| 65 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
0.743 |
-0.287 |
| 66 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.741 |
0.843 |
| 67 |
263032_at |
AT1G23850
|
unknown |
0.737 |
-0.014 |
| 68 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
0.734 |
-0.161 |
| 69 |
252097_at |
AT3G51090
|
unknown |
0.732 |
-0.279 |
| 70 |
257522_at |
AT3G08990
|
[Yippee family putative zinc-binding protein] |
0.732 |
0.178 |
| 71 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.731 |
0.344 |
| 72 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.726 |
-0.366 |
| 73 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.723 |
-0.529 |
| 74 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.722 |
0.456 |
| 75 |
245076_at |
AT2G23170
|
GH3.3 |
0.722 |
0.478 |
| 76 |
265620_at |
AT2G27310
|
[F-box family protein] |
0.721 |
0.441 |
| 77 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.720 |
0.167 |
| 78 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.720 |
0.074 |
| 79 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.713 |
0.459 |
| 80 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.713 |
0.651 |
| 81 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.713 |
0.676 |
| 82 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.711 |
-0.138 |
| 83 |
256349_at |
AT1G54890
|
[Late embryogenesis abundant (LEA) protein-related] |
0.710 |
-0.147 |
| 84 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.707 |
0.633 |
| 85 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.703 |
0.704 |
| 86 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
0.701 |
1.291 |
| 87 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
0.701 |
0.177 |
| 88 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.700 |
0.866 |
| 89 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.698 |
0.155 |
| 90 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.698 |
0.099 |
| 91 |
246222_at |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
0.697 |
0.505 |
| 92 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.696 |
0.017 |
| 93 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.696 |
0.137 |
| 94 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.690 |
1.120 |
| 95 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
0.689 |
-0.246 |
| 96 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.686 |
-0.173 |
| 97 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.684 |
0.687 |
| 98 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.682 |
0.637 |
| 99 |
258652_at |
AT3G09910
|
ATRAB18C, ATRABC2B, RAB GTPase homolog C2B, RABC2b, RAB GTPase homolog C2B |
0.680 |
-0.006 |
| 100 |
260204_at |
AT1G52900
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.679 |
0.166 |
| 101 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.678 |
-0.186 |
| 102 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.676 |
0.512 |
| 103 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.675 |
0.333 |
| 104 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.674 |
0.261 |
| 105 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.670 |
0.398 |
| 106 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.663 |
0.183 |
| 107 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.663 |
0.379 |
| 108 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.661 |
0.391 |
| 109 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
0.661 |
-0.048 |
| 110 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.654 |
0.109 |
| 111 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.653 |
-0.271 |
| 112 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.653 |
0.030 |
| 113 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
0.650 |
0.229 |
| 114 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.650 |
1.125 |
| 115 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.649 |
-0.070 |
| 116 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.647 |
0.486 |
| 117 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.645 |
0.103 |
| 118 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.643 |
0.527 |
| 119 |
261748_at |
AT1G76070
|
unknown |
0.643 |
0.587 |
| 120 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.643 |
-0.315 |
| 121 |
264882_at |
AT1G61110
|
anac025, NAC domain containing protein 25, NAC025, NAC domain containing protein 25 |
0.639 |
0.180 |
| 122 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
0.639 |
0.040 |
| 123 |
266452_at |
AT2G43320
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.634 |
0.229 |
| 124 |
251132_at |
AT5G01200
|
[Duplicated homeodomain-like superfamily protein] |
0.634 |
0.235 |
| 125 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.633 |
-0.034 |
| 126 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.633 |
0.351 |
| 127 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
0.632 |
0.167 |
| 128 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.632 |
-0.118 |
| 129 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.630 |
0.021 |
| 130 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.629 |
0.289 |
| 131 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.628 |
0.654 |
| 132 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.625 |
0.054 |
| 133 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.625 |
0.093 |
| 134 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.625 |
0.634 |
| 135 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.622 |
0.334 |
| 136 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.618 |
0.266 |
| 137 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.616 |
0.780 |
| 138 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.615 |
1.468 |
| 139 |
260841_at |
AT1G29195
|
unknown |
0.614 |
-0.238 |
| 140 |
255527_at |
AT4G02360
|
unknown |
0.614 |
-0.089 |
| 141 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
0.614 |
-0.059 |
| 142 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.613 |
0.212 |
| 143 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.611 |
0.049 |
| 144 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.611 |
-0.371 |
| 145 |
249406_at |
AT5G40210
|
UMAMIT42, Usually multiple acids move in and out Transporters 42 |
0.610 |
0.116 |
| 146 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.610 |
0.022 |
| 147 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.609 |
0.448 |
| 148 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.608 |
0.062 |
| 149 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.606 |
0.013 |
| 150 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.606 |
1.054 |
| 151 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.603 |
0.076 |
| 152 |
255900_at |
AT1G17830
|
unknown |
0.603 |
-0.109 |
| 153 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.600 |
0.160 |
| 154 |
247287_at |
AT5G64230
|
unknown |
0.600 |
0.571 |
| 155 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
0.600 |
0.245 |
| 156 |
246584_at |
AT5G14730
|
unknown |
0.600 |
0.959 |
| 157 |
250918_at |
AT5G03610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.598 |
-0.241 |
| 158 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
0.597 |
0.451 |
| 159 |
253827_at |
AT4G28085
|
unknown |
0.597 |
-0.174 |
| 160 |
259725_at |
AT1G61065
|
unknown |
0.596 |
0.189 |
| 161 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.593 |
0.345 |
| 162 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
0.591 |
0.252 |
| 163 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.590 |
-0.144 |
| 164 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.586 |
0.664 |
| 165 |
257517_at |
AT3G16330
|
unknown |
0.585 |
0.253 |
| 166 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
0.584 |
-0.257 |
| 167 |
252003_at |
AT3G52770
|
ZPR3, LITTLE ZIPPER 3 |
0.581 |
0.070 |
| 168 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.581 |
0.278 |
| 169 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.580 |
0.110 |
| 170 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.579 |
0.026 |
| 171 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.579 |
-0.108 |
| 172 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
0.577 |
-0.284 |
| 173 |
249379_at |
AT5G40460
|
unknown |
0.576 |
0.235 |
| 174 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.574 |
0.867 |
| 175 |
253124_at |
AT4G36030
|
ARO3, armadillo repeat only 3 |
0.573 |
0.168 |
| 176 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.572 |
0.714 |
| 177 |
255728_at |
AT1G25500
|
[Plasma-membrane choline transporter family protein] |
0.572 |
-0.187 |
| 178 |
261455_at |
AT1G21070
|
[Nucleotide-sugar transporter family protein] |
0.571 |
-0.315 |
| 179 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.569 |
0.129 |
| 180 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.568 |
0.151 |
| 181 |
255132_at |
AT4G08170
|
[Inositol 1,3,4-trisphosphate 5/6-kinase family protein] |
0.568 |
0.260 |
| 182 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
0.566 |
0.215 |
| 183 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.566 |
0.091 |
| 184 |
250580_at |
AT5G07440
|
GDH2, glutamate dehydrogenase 2 |
0.565 |
0.187 |
| 185 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.565 |
0.290 |
| 186 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.559 |
0.317 |
| 187 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.558 |
0.417 |
| 188 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.557 |
-0.020 |
| 189 |
263151_at |
AT1G54120
|
unknown |
0.555 |
-0.162 |
| 190 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.555 |
0.219 |
| 191 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.555 |
0.461 |
| 192 |
249134_at |
AT5G43150
|
unknown |
0.555 |
0.035 |
| 193 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.555 |
0.190 |
| 194 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.550 |
0.012 |
| 195 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.549 |
-0.088 |
| 196 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
0.549 |
0.059 |
| 197 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.548 |
-0.212 |
| 198 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.548 |
-0.267 |
| 199 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.546 |
0.507 |
| 200 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.545 |
0.026 |
| 201 |
255878_at |
AT2G40620
|
[Basic-leucine zipper (bZIP) transcription factor family protein] |
0.545 |
0.046 |
| 202 |
254597_at |
AT4G18980
|
AtS40-3, AtS40-3 |
0.544 |
0.155 |
| 203 |
245353_at |
AT4G16000
|
unknown |
0.543 |
0.046 |
| 204 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
0.542 |
0.318 |
| 205 |
246779_at |
AT5G27520
|
AtPNC2, PNC2, peroxisomal adenine nucleotide carrier 2 |
0.542 |
0.185 |
| 206 |
248592_at |
AT5G49280
|
[hydroxyproline-rich glycoprotein family protein] |
0.541 |
-0.047 |
| 207 |
265648_at |
AT2G27500
|
[Glycosyl hydrolase superfamily protein] |
0.541 |
0.229 |
| 208 |
247047_at |
AT5G66650
|
unknown |
0.541 |
-0.002 |
| 209 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
0.541 |
-0.087 |
| 210 |
258207_at |
AT3G14050
|
AT-RSH2, RELA-SPOT HOMOLOG 2, ATRSH2, RELA/SPOT HOMOLOG 2, RSH2, RELA/SPOT homolog 2 |
0.540 |
0.183 |
| 211 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.540 |
0.523 |
| 212 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
0.537 |
0.190 |
| 213 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.535 |
0.409 |
| 214 |
253274_at |
AT4G34200
|
EDA9, embryo sac development arrest 9, PGDH1, phosphoglycerate dehydrogenase 1 |
0.535 |
-0.037 |
| 215 |
247922_at |
AT5G57500
|
[Galactosyltransferase family protein] |
0.535 |
0.057 |
| 216 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
0.535 |
-0.009 |
| 217 |
259057_at |
AT3G03310
|
ATLCAT3, ARABIDOPSIS LECITHIN:CHOLESTEROL ACYLTRANSFERASE 3, LCAT3, lecithin:cholesterol acyltransferase 3 |
0.533 |
0.518 |
| 218 |
256037_at |
AT1G19160
|
[F-box family protein] |
0.530 |
0.133 |
| 219 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.530 |
0.014 |
| 220 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.529 |
0.578 |
| 221 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.526 |
0.184 |
| 222 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.525 |
0.114 |
| 223 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.525 |
-0.070 |
| 224 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.525 |
0.844 |
| 225 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.524 |
0.317 |
| 226 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
0.524 |
0.365 |
| 227 |
258551_at |
AT3G06890
|
unknown |
0.522 |
-0.081 |
| 228 |
251207_at |
AT3G63050
|
unknown |
0.521 |
0.051 |
| 229 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
0.519 |
0.262 |
| 230 |
254293_at |
AT4G23060
|
IQD22, IQ-domain 22 |
0.519 |
-0.093 |
| 231 |
248980_at |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
0.519 |
0.179 |
| 232 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.518 |
0.489 |
| 233 |
264543_at |
AT1G55790
|
[FUNCTIONS IN: metal ion binding] |
0.518 |
0.160 |
| 234 |
263475_at |
AT2G31945
|
unknown |
0.518 |
0.602 |
| 235 |
253919_at |
AT4G27350
|
unknown |
0.517 |
0.565 |
| 236 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
0.517 |
0.456 |
| 237 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.516 |
0.030 |
| 238 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.511 |
0.202 |
| 239 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
0.511 |
-0.076 |
| 240 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.511 |
-0.347 |
| 241 |
267058_at |
AT2G32510
|
MAPKKK17, mitogen-activated protein kinase kinase kinase 17 |
0.510 |
0.884 |
| 242 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
0.510 |
0.336 |
| 243 |
257226_at |
AT3G27880
|
unknown |
0.510 |
0.512 |
| 244 |
251023_at |
AT5G02170
|
[Transmembrane amino acid transporter family protein] |
0.509 |
0.276 |
| 245 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.508 |
0.474 |
| 246 |
245755_at |
AT1G35210
|
unknown |
0.506 |
0.183 |
| 247 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.506 |
-0.132 |
| 248 |
250158_at |
AT5G15190
|
unknown |
0.505 |
0.330 |
| 249 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
0.504 |
0.084 |
| 250 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
0.503 |
-0.024 |
| 251 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.503 |
0.306 |
| 252 |
259749_at |
AT1G71100
|
RSW10, RADIAL SWELLING 10 |
0.503 |
0.339 |
| 253 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.502 |
0.090 |
| 254 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.501 |
-0.071 |
| 255 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.499 |
0.218 |
| 256 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.498 |
-0.061 |
| 257 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
0.497 |
0.937 |
| 258 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.497 |
0.001 |
| 259 |
259400_at |
AT1G17750
|
AtPEPR2, PEP1 RECEPTOR 2, PEPR2, PEP1 receptor 2 |
0.496 |
-0.075 |
| 260 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
0.496 |
-0.204 |
| 261 |
247193_at |
AT5G65380
|
[MATE efflux family protein] |
0.496 |
0.116 |
| 262 |
260243_at |
AT1G63720
|
unknown |
0.496 |
0.391 |
| 263 |
265913_at |
AT2G25625
|
unknown |
0.495 |
0.263 |
| 264 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.494 |
0.341 |
| 265 |
256793_at |
AT3G22160
|
JAV1, jasmonate-associated VQ motif gene 1 |
0.493 |
0.082 |
| 266 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.491 |
0.144 |
| 267 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.490 |
0.302 |
| 268 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.489 |
0.444 |
| 269 |
249042_at |
AT5G44350
|
[ethylene-responsive nuclear protein -related] |
0.489 |
0.106 |
| 270 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.489 |
-0.199 |
| 271 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
0.488 |
-0.131 |
| 272 |
247412_at |
AT5G63010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.488 |
0.048 |
| 273 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.487 |
0.111 |
| 274 |
246842_at |
AT5G26731
|
unknown |
0.486 |
-0.526 |
| 275 |
260179_at |
AT1G70690
|
HWI1, HOPW1-1-INDUCED GENE1, PDLP5, PLASMODESMATA-LOCATED PROTEIN 5 |
0.485 |
-0.184 |
| 276 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.485 |
0.472 |
| 277 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.483 |
0.088 |
| 278 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.483 |
0.388 |
| 279 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.482 |
-0.150 |
| 280 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.482 |
0.161 |
| 281 |
261973_at |
AT1G64610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.481 |
-0.024 |
| 282 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.480 |
0.003 |
| 283 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.479 |
0.646 |
| 284 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.479 |
0.337 |
| 285 |
245545_at |
AT4G15280
|
UGT71B5, UDP-glucosyl transferase 71B5 |
0.479 |
0.220 |
| 286 |
247333_at |
AT5G63600
|
ATFLS5, FLS5, flavonol synthase 5 |
0.479 |
0.046 |
| 287 |
258209_at |
AT3G14060
|
unknown |
0.479 |
0.218 |
| 288 |
266195_at |
AT2G39100
|
[RING/U-box superfamily protein] |
0.478 |
0.330 |
| 289 |
264660_at |
AT1G09940
|
HEMA2 |
0.478 |
0.434 |
| 290 |
250267_at |
AT5G12930
|
unknown |
0.478 |
-0.055 |
| 291 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.477 |
0.762 |
| 292 |
256725_at |
AT2G34070
|
TBL37, TRICHOME BIREFRINGENCE-LIKE 37 |
0.477 |
0.074 |
| 293 |
265539_at |
AT2G15830
|
unknown |
0.474 |
-0.277 |
| 294 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.473 |
-0.014 |
| 295 |
250161_at |
AT5G15240
|
[Transmembrane amino acid transporter family protein] |
0.472 |
0.010 |
| 296 |
251061_at |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
0.472 |
0.194 |
| 297 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.470 |
0.600 |
| 298 |
251642_at |
AT3G57520
|
AtSIP2, seed imbibition 2, RS2, raffinose synthase 2, SIP2, seed imbibition 2 |
0.470 |
0.407 |
| 299 |
262183_at |
AT1G77890
|
[DNA-directed RNA polymerase II protein] |
0.470 |
0.236 |
| 300 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.470 |
-0.244 |
| 301 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-1.101 |
-0.097 |
| 302 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-1.098 |
0.060 |
| 303 |
262598_at |
AT1G15260
|
unknown |
-1.000 |
-0.765 |
| 304 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.976 |
0.011 |
| 305 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.964 |
0.006 |
| 306 |
248186_at |
AT5G53880
|
unknown |
-0.956 |
-0.028 |
| 307 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.954 |
-0.109 |
| 308 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.936 |
-0.552 |
| 309 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.918 |
-0.341 |
| 310 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.909 |
-0.025 |
| 311 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.906 |
-0.321 |
| 312 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.902 |
-0.362 |
| 313 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.868 |
-0.349 |
| 314 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.866 |
0.131 |
| 315 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.865 |
-0.222 |
| 316 |
257974_at |
AT3G20820
|
[Leucine-rich repeat (LRR) family protein] |
-0.860 |
-0.300 |
| 317 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.846 |
0.252 |
| 318 |
252353_at |
AT3G48200
|
unknown |
-0.843 |
-0.180 |
| 319 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.834 |
-0.480 |
| 320 |
258804_at |
AT3G04760
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.834 |
-0.476 |
| 321 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.831 |
-0.224 |
| 322 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.828 |
-0.626 |
| 323 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.822 |
-0.480 |
| 324 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.821 |
0.144 |
| 325 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.814 |
0.173 |
| 326 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.811 |
0.130 |
| 327 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.809 |
-0.296 |
| 328 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.809 |
-0.531 |
| 329 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.809 |
0.150 |
| 330 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.808 |
0.015 |
| 331 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.793 |
-0.285 |
| 332 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
-0.791 |
-0.013 |
| 333 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.780 |
-0.367 |
| 334 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.779 |
-0.300 |
| 335 |
264201_at |
AT1G22630
|
unknown |
-0.776 |
-0.192 |
| 336 |
258468_at |
AT3G06070
|
unknown |
-0.774 |
0.273 |
| 337 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.765 |
-0.425 |
| 338 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.764 |
-0.405 |
| 339 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.764 |
-0.422 |
| 340 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
-0.763 |
-0.460 |
| 341 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.760 |
-0.289 |
| 342 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.759 |
-0.539 |
| 343 |
245906_at |
AT5G11070
|
unknown |
-0.759 |
0.082 |
| 344 |
263597_at |
AT2G01870
|
unknown |
-0.756 |
-0.560 |
| 345 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.756 |
-0.318 |
| 346 |
247977_at |
AT5G56850
|
unknown |
-0.755 |
-0.075 |
| 347 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.752 |
-0.391 |
| 348 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.751 |
0.025 |
| 349 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.748 |
0.126 |
| 350 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.742 |
0.112 |
| 351 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.739 |
-0.216 |
| 352 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.738 |
-0.078 |
| 353 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
-0.734 |
0.037 |
| 354 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.732 |
0.284 |
| 355 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.721 |
-0.113 |
| 356 |
264930_at |
AT1G60800
|
AtNIK3, NIK3, NSP-interacting kinase 3 |
-0.718 |
-0.261 |
| 357 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.717 |
-0.048 |
| 358 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.716 |
-0.013 |
| 359 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.716 |
-0.050 |
| 360 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.716 |
-0.270 |
| 361 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.712 |
-0.539 |
| 362 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.704 |
-0.086 |
| 363 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.701 |
-0.119 |
| 364 |
245165_at |
AT2G33180
|
unknown |
-0.700 |
-0.523 |
| 365 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.699 |
-0.339 |
| 366 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.698 |
-0.506 |
| 367 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.698 |
-0.531 |
| 368 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.696 |
-0.001 |
| 369 |
247853_at |
AT5G58140
|
NPL1, NON PHOTOTROPIC HYPOCOTYL 1-LIKE, PHOT2, phototropin 2 |
-0.695 |
0.112 |
| 370 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.693 |
-0.000 |
| 371 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.691 |
-0.018 |
| 372 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.689 |
-0.376 |
| 373 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
-0.687 |
0.110 |
| 374 |
264958_at |
AT1G76960
|
unknown |
-0.685 |
0.406 |
| 375 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.685 |
-0.082 |
| 376 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.683 |
0.226 |
| 377 |
263846_at |
AT2G36990
|
ATSIG6, SIGMA FACTOR 6, SIG6, SIGMA FACTOR 6, SIGF, RNApolymerase sigma-subunit F, SOLDAT8 |
-0.682 |
-0.153 |
| 378 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.682 |
-0.383 |
| 379 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.682 |
-0.329 |
| 380 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.680 |
0.193 |
| 381 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.678 |
-0.219 |
| 382 |
248095_at |
AT5G55230
|
ATMAP65-1, microtubule-associated proteins 65-1, MAP65-1, MAP65-1, microtubule-associated proteins 65-1 |
-0.675 |
0.026 |
| 383 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.675 |
-0.106 |
| 384 |
262693_at |
AT1G62780
|
unknown |
-0.675 |
-0.263 |
| 385 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.675 |
-0.102 |
| 386 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.674 |
-0.244 |
| 387 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.673 |
0.136 |
| 388 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.672 |
0.062 |
| 389 |
263674_at |
AT2G04790
|
unknown |
-0.669 |
-0.180 |
| 390 |
265990_at |
AT2G24280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.666 |
-0.251 |
| 391 |
265993_at |
AT2G24160
|
[pseudogene] |
-0.665 |
0.123 |
| 392 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.663 |
0.117 |
| 393 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.662 |
-0.310 |
| 394 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
-0.659 |
-0.263 |
| 395 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.656 |
0.084 |
| 396 |
259460_at |
AT1G44000
|
unknown |
-0.654 |
-0.167 |
| 397 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.652 |
-0.113 |
| 398 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.652 |
-0.286 |
| 399 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.649 |
-0.206 |
| 400 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.646 |
0.087 |
| 401 |
247288_at |
AT5G64330
|
JK218, NPH3, NON-PHOTOTROPIC HYPOCOTYL 3, RPT3, ROOT PHOTOTROPISM 3 |
-0.645 |
0.271 |
| 402 |
247383_at |
AT5G63410
|
[Leucine-rich repeat protein kinase family protein] |
-0.643 |
-0.217 |
| 403 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.643 |
-0.096 |
| 404 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.642 |
-0.170 |
| 405 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.640 |
-0.253 |
| 406 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.640 |
-0.147 |
| 407 |
248329_at |
AT5G52780
|
unknown |
-0.639 |
-0.538 |
| 408 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.636 |
-0.103 |
| 409 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.636 |
-0.310 |
| 410 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
-0.631 |
-0.354 |
| 411 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.629 |
-0.068 |
| 412 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.628 |
-0.480 |
| 413 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.625 |
0.136 |
| 414 |
254737_at |
AT4G13840
|
CER26, ECERIFERUM 26 |
-0.625 |
-0.175 |
| 415 |
256796_at |
AT3G22210
|
unknown |
-0.624 |
-0.860 |
| 416 |
258156_at |
AT3G18050
|
unknown |
-0.623 |
-0.090 |
| 417 |
248920_at |
AT5G45930
|
CHL I2, CHLI-2, CHLI2, magnesium chelatase i2 |
-0.620 |
-0.398 |
| 418 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.620 |
-0.045 |
| 419 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.617 |
-0.042 |
| 420 |
259179_at |
AT3G01660
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.617 |
-0.373 |
| 421 |
253758_at |
AT4G29060
|
emb2726, embryo defective 2726 |
-0.617 |
-0.207 |
| 422 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.615 |
-0.245 |
| 423 |
262286_at |
AT1G68585
|
unknown |
-0.614 |
0.357 |
| 424 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.614 |
-0.451 |
| 425 |
259258_at |
AT3G07670
|
[Rubisco methyltransferase family protein] |
-0.613 |
-0.215 |
| 426 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.612 |
0.015 |
| 427 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.612 |
0.001 |
| 428 |
246231_at |
AT4G37080
|
unknown |
-0.611 |
-0.383 |
| 429 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
-0.610 |
-0.165 |
| 430 |
259207_at |
AT3G09050
|
unknown |
-0.609 |
-0.302 |
| 431 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.608 |
0.254 |
| 432 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.607 |
-0.033 |
| 433 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.607 |
-0.581 |
| 434 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.605 |
0.163 |
| 435 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.604 |
-0.280 |
| 436 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.602 |
-0.127 |
| 437 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.601 |
-0.322 |
| 438 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.599 |
-0.376 |
| 439 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.599 |
-0.588 |
| 440 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.598 |
-0.131 |
| 441 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.597 |
-0.521 |
| 442 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.595 |
-0.286 |
| 443 |
250429_at |
AT5G10470
|
KAC1, kinesin like protein for actin based chloroplast movement 1, KCA1, KINESIN CDKA;1 ASSOCIATED 1 |
-0.595 |
-0.148 |
| 444 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.595 |
-0.269 |
| 445 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.595 |
-0.071 |
| 446 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
-0.592 |
-0.242 |
| 447 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.591 |
0.107 |
| 448 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.589 |
0.036 |
| 449 |
263915_at |
AT2G36430
|
unknown |
-0.589 |
-0.083 |
| 450 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
-0.589 |
-0.703 |
| 451 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.587 |
0.517 |
| 452 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.587 |
0.139 |
| 453 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.585 |
0.035 |
| 454 |
262648_at |
AT1G14030
|
LSMT-L, lysine methyltransferase (LSMT)-like |
-0.585 |
-0.265 |
| 455 |
252001_at |
AT3G52750
|
FTSZ2-2 |
-0.584 |
-0.053 |
| 456 |
260347_at |
AT1G69420
|
[DHHC-type zinc finger family protein] |
-0.584 |
-0.103 |
| 457 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.583 |
-0.377 |
| 458 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.583 |
-0.046 |
| 459 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
-0.582 |
-0.172 |
| 460 |
253903_at |
AT4G27180
|
ATK2, kinesin 2, KATB, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA B |
-0.581 |
0.005 |
| 461 |
257647_at |
AT3G25805
|
unknown |
-0.579 |
-0.054 |
| 462 |
253283_at |
AT4G34090
|
unknown |
-0.579 |
-0.225 |
| 463 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.578 |
-0.385 |
| 464 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.578 |
0.055 |
| 465 |
261500_at |
AT1G28400
|
unknown |
-0.577 |
-0.627 |
| 466 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
-0.577 |
-0.046 |
| 467 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.577 |
-0.199 |
| 468 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.577 |
-0.294 |
| 469 |
263987_at |
AT2G42690
|
[alpha/beta-Hydrolases superfamily protein] |
-0.572 |
-0.101 |
| 470 |
247232_at |
AT5G64940
|
ATATH13, ARABIDOPSIS THALIANA ABC2 HOMOLOG 13, ATH13, ABC2 homolog 13, ATOSA1, A. THALIANA OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1, OSA1, OXIDATIVE STRESS-RELATED ABC1-LIKE PROTEIN 1 |
-0.572 |
-0.009 |
| 471 |
262283_at |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
-0.572 |
-0.303 |
| 472 |
262397_at |
AT1G49380
|
[cytochrome c biogenesis protein family] |
-0.571 |
-0.270 |
| 473 |
245952_at |
AT5G28500
|
unknown |
-0.571 |
-0.334 |
| 474 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.570 |
-0.082 |
| 475 |
245347_at |
AT4G14890
|
FdC1, ferredoxin C 1 |
-0.569 |
-0.378 |
| 476 |
249900_at |
AT5G22640
|
emb1211, embryo defective 1211, TIC100, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 100 |
-0.568 |
-0.180 |
| 477 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.567 |
-0.513 |
| 478 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.562 |
-0.182 |
| 479 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.561 |
0.067 |
| 480 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.560 |
-0.054 |
| 481 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.560 |
0.056 |
| 482 |
257773_at |
AT3G29185
|
unknown |
-0.559 |
-0.144 |
| 483 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.556 |
0.092 |
| 484 |
246901_at |
AT5G25630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.555 |
0.022 |
| 485 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.555 |
-0.245 |
| 486 |
267637_at |
AT2G42190
|
unknown |
-0.555 |
-0.326 |
| 487 |
263442_at |
AT2G28605
|
[Photosystem II reaction center PsbP family protein] |
-0.554 |
-0.390 |
| 488 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.553 |
-0.515 |
| 489 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
-0.553 |
-0.081 |
| 490 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
-0.551 |
-0.207 |
| 491 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.550 |
-0.231 |
| 492 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.548 |
-0.174 |
| 493 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.548 |
-0.053 |
| 494 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.547 |
-0.210 |
| 495 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
-0.545 |
-0.105 |
| 496 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.544 |
-0.088 |
| 497 |
266575_at |
AT2G24060
|
[Translation initiation factor 3 protein] |
-0.542 |
-0.169 |
| 498 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.542 |
0.008 |
| 499 |
249007_at |
AT5G44650
|
AtCEST, Arabidopsis thaliana chloroplast protein-enhancing stress tolerance, CEST, chloroplast protein-enhancing stress tolerance, Y3IP1, Ycf3-interacting protein 1 |
-0.540 |
-0.275 |
| 500 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.540 |
0.039 |
| 501 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.539 |
0.464 |
| 502 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.534 |
0.027 |
| 503 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
-0.533 |
0.017 |
| 504 |
261498_at |
AT1G28440
|
HSL1, HAESA-like 1 |
-0.532 |
-0.225 |
| 505 |
263696_at |
AT1G31230
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH I, aspartate kinase-homoserine dehydrogenase i |
-0.531 |
-0.269 |
| 506 |
255720_at |
AT1G32060
|
PRK, phosphoribulokinase |
-0.529 |
-0.069 |
| 507 |
266929_at |
AT2G45850
|
AHL9, AT-hook motif nuclear localized protein 9 |
-0.529 |
-0.244 |
| 508 |
252277_at |
AT3G49470
|
NACA2, nascent polypeptide-associated complex subunit alpha-like protein 2 |
-0.528 |
-0.013 |
| 509 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.525 |
0.136 |
| 510 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.525 |
-0.585 |
| 511 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.524 |
-0.051 |
| 512 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.523 |
-0.211 |
| 513 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.523 |
-0.395 |
| 514 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.522 |
-0.028 |
| 515 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
-0.521 |
-0.247 |
| 516 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.521 |
0.177 |
| 517 |
256115_at |
AT1G16880
|
ACR11, ACT domain repeats 11 |
-0.517 |
-0.059 |
| 518 |
245492_at |
AT4G16340
|
SPK1, SPIKE1 |
-0.517 |
-0.241 |
| 519 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.517 |
-0.201 |
| 520 |
248046_at |
AT5G56040
|
SKM2, STERILITY-REGULATING KINASE MEMBER 2 |
-0.516 |
-0.232 |
| 521 |
262377_at |
AT1G73110
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.515 |
-0.132 |
| 522 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
-0.515 |
-0.366 |
| 523 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.514 |
0.103 |
| 524 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.512 |
-0.167 |
| 525 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
-0.512 |
-0.105 |
| 526 |
266990_at |
AT2G39190
|
ATATH8 |
-0.512 |
-0.040 |
| 527 |
253548_at |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.511 |
-0.146 |
| 528 |
258566_at |
AT3G04110
|
ATGLR1.1, GLR1, GLUTAMATE RECEPTOR 1, GLR1.1, glutamate receptor 1.1 |
-0.511 |
-0.022 |
| 529 |
257510_at |
AT1G55360
|
unknown |
-0.511 |
-0.154 |
| 530 |
264096_at |
AT1G78995
|
unknown |
-0.508 |
-0.082 |
| 531 |
254623_at |
AT4G18480
|
CH-42, CHLORINA 42, CH42, CHLORINA 42, CHL11, CHLI-1, CHLI1 |
-0.508 |
-0.240 |
| 532 |
245845_at |
AT1G26150
|
AtPERK10, proline-rich extensin-like receptor kinase 10, PERK10, proline-rich extensin-like receptor kinase 10 |
-0.508 |
-0.073 |
| 533 |
247850_at |
AT5G58150
|
[Leucine-rich repeat protein kinase family protein] |
-0.506 |
-0.199 |
| 534 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.505 |
-0.254 |
| 535 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.505 |
-0.132 |
| 536 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.505 |
-0.018 |
| 537 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.504 |
0.160 |
| 538 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
-0.504 |
-0.397 |
| 539 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.503 |
0.030 |
| 540 |
256383_at |
AT1G66820
|
[glycine-rich protein] |
-0.503 |
-0.287 |
| 541 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.501 |
-0.087 |
| 542 |
264394_at |
AT1G11860
|
[Glycine cleavage T-protein family] |
-0.501 |
-0.027 |
| 543 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.501 |
-0.116 |
| 544 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.500 |
-0.130 |
| 545 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.500 |
0.094 |
| 546 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.500 |
-0.152 |
| 547 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
-0.497 |
0.233 |
| 548 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.496 |
-0.067 |
| 549 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.496 |
0.021 |
| 550 |
263410_at |
AT2G04039
|
unknown |
-0.495 |
-0.196 |
| 551 |
265967_at |
AT2G37450
|
UMAMIT13, Usually multiple acids move in and out Transporters 13 |
-0.493 |
-0.133 |
| 552 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.493 |
-0.244 |
| 553 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.492 |
0.079 |
| 554 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
-0.492 |
-0.278 |
| 555 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.492 |
0.035 |
| 556 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.492 |
0.122 |
| 557 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.492 |
0.262 |
| 558 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.491 |
-0.079 |
| 559 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.490 |
-0.409 |
| 560 |
261197_at |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
-0.490 |
-0.143 |
| 561 |
258324_at |
AT3G22780
|
ATTSO1, TSO1, CHINESE FOR 'UGLY' |
-0.490 |
-0.129 |
| 562 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.489 |
-0.371 |
| 563 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.488 |
-0.035 |
| 564 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
-0.488 |
-0.205 |
| 565 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.488 |
0.018 |
| 566 |
256622_at |
AT3G28920
|
AtHB34, homeobox protein 34, HB34, homeobox protein 34, ZHD9, ZINC FINGER HOMEODOMAIN 9 |
-0.487 |
-0.325 |
| 567 |
247192_at |
AT5G65360
|
H3.1, histone 3.1 |
-0.486 |
-0.291 |
| 568 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.484 |
0.206 |
| 569 |
252561_at |
AT3G45980
|
H2B, HISTONE H2B, HTB9 |
-0.484 |
-0.084 |
| 570 |
246701_at |
AT5G28020
|
ATCYSD2, CYSTEINE SYNTHASE D2, CYSD2, cysteine synthase D2 |
-0.483 |
0.112 |
| 571 |
261375_at |
AT1G53160
|
FTM6, FLORAL TRANSITION AT THE MERISTEM6, SPL4, squamosa promoter binding protein-like 4 |
-0.482 |
0.126 |
| 572 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
-0.482 |
-0.047 |
| 573 |
255957_at |
AT1G22160
|
unknown |
-0.482 |
0.215 |
| 574 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.482 |
-0.330 |
| 575 |
254815_at |
AT4G12420
|
SKU5 |
-0.482 |
-0.017 |
| 576 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.481 |
-0.271 |
| 577 |
252033_at |
AT3G51950
|
[Zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein] |
-0.480 |
-0.084 |
| 578 |
246792_at |
AT5G27290
|
unknown |
-0.480 |
-0.041 |
| 579 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.480 |
0.107 |
| 580 |
247891_at |
AT5G57960
|
[GTP-binding protein, HflX] |
-0.479 |
-0.159 |
| 581 |
255440_at |
AT4G02530
|
[chloroplast thylakoid lumen protein] |
-0.479 |
-0.118 |
| 582 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.478 |
-0.078 |
| 583 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.476 |
-0.651 |
| 584 |
253424_at |
AT4G32330
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.475 |
-0.152 |
| 585 |
246487_at |
AT5G16030
|
unknown |
-0.474 |
-0.402 |
| 586 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
-0.474 |
0.016 |
| 587 |
246929_at |
AT5G25210
|
unknown |
-0.474 |
-0.251 |
| 588 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.473 |
0.157 |
| 589 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.473 |
-0.169 |
| 590 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.472 |
-0.179 |
| 591 |
251936_at |
AT3G53700
|
MEE40, maternal effect embryo arrest 40 |
-0.471 |
-0.274 |
| 592 |
250498_at |
AT5G09660
|
PMDH2, peroxisomal NAD-malate dehydrogenase 2 |
-0.469 |
0.035 |
| 593 |
257205_at |
AT3G16520
|
UGT88A1, UDP-glucosyl transferase 88A1 |
-0.469 |
-0.078 |
| 594 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
-0.468 |
-0.059 |
| 595 |
262483_at |
AT1G17220
|
FUG1, fu-gaeri1 |
-0.468 |
-0.028 |
| 596 |
252130_at |
AT3G50820
|
OEC33, OXYGEN EVOLVING COMPLEX SUBUNIT 33 KDA, PSBO-2, PHOTOSYSTEM II SUBUNIT O-2, PSBO2, photosystem II subunit O-2 |
-0.468 |
0.145 |
| 597 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.467 |
0.536 |
| 598 |
250739_at |
AT5G05740
|
ATEGY2, EGY2, ethylene-dependent gravitropism-deficient and yellow-green-like 2 |
-0.465 |
-0.178 |
| 599 |
248224_at |
AT5G53490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.465 |
-0.259 |
| 600 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.464 |
-0.193 |