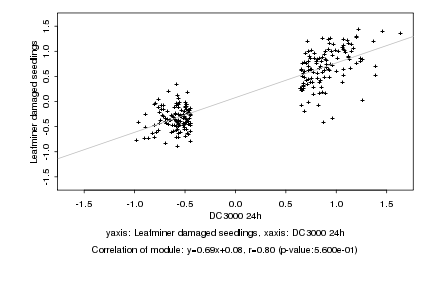
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
DC3000 24h |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
1.633 |
1.360 |
| 2 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
1.456 |
1.403 |
| 3 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
1.382 |
0.529 |
| 4 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
1.380 |
0.699 |
| 5 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
1.361 |
1.212 |
| 6 |
253070_at |
AT4G37850
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.257 |
0.037 |
| 7 |
259054_at |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
1.257 |
0.846 |
| 8 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
1.238 |
0.807 |
| 9 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
1.231 |
0.872 |
| 10 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.215 |
1.437 |
| 11 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
1.198 |
1.277 |
| 12 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.193 |
1.298 |
| 13 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
1.192 |
0.758 |
| 14 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
1.165 |
1.068 |
| 15 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.149 |
1.010 |
| 16 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.147 |
1.152 |
| 17 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
1.137 |
0.671 |
| 18 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.128 |
0.843 |
| 19 |
245209_at |
AT5G12340
|
unknown |
1.120 |
1.171 |
| 20 |
245443_at |
AT4G16730
|
TPS02, terpene synthase 02 |
1.119 |
0.883 |
| 21 |
255527_at |
AT4G02360
|
unknown |
1.117 |
0.911 |
| 22 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
1.104 |
1.221 |
| 23 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
1.082 |
0.981 |
| 24 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
1.075 |
1.035 |
| 25 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
1.069 |
1.117 |
| 26 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.069 |
1.244 |
| 27 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
1.067 |
1.044 |
| 28 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
1.067 |
0.531 |
| 29 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
1.066 |
0.651 |
| 30 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
1.058 |
0.381 |
| 31 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
1.056 |
1.085 |
| 32 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.022 |
1.015 |
| 33 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
1.008 |
0.865 |
| 34 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.999 |
0.609 |
| 35 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.984 |
1.034 |
| 36 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.968 |
1.150 |
| 37 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.961 |
0.929 |
| 38 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
0.960 |
0.727 |
| 39 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.955 |
-0.334 |
| 40 |
267206_at |
AT2G30830
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.946 |
0.998 |
| 41 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.938 |
1.262 |
| 42 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.937 |
0.744 |
| 43 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.926 |
1.160 |
| 44 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.926 |
0.487 |
| 45 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.924 |
0.621 |
| 46 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.918 |
0.654 |
| 47 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.918 |
1.239 |
| 48 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.915 |
1.038 |
| 49 |
251633_at |
AT3G57460
|
[catalytics] |
0.899 |
0.679 |
| 50 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.895 |
0.749 |
| 51 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.894 |
0.820 |
| 52 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.892 |
0.162 |
| 53 |
247047_at |
AT5G66650
|
unknown |
0.888 |
0.999 |
| 54 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.886 |
0.853 |
| 55 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
0.882 |
0.491 |
| 56 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
0.880 |
0.629 |
| 57 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.875 |
0.951 |
| 58 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.873 |
-0.402 |
| 59 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.858 |
0.192 |
| 60 |
263032_at |
AT1G23850
|
unknown |
0.856 |
0.580 |
| 61 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.854 |
0.877 |
| 62 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.854 |
1.263 |
| 63 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.847 |
0.291 |
| 64 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.845 |
0.814 |
| 65 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.842 |
0.716 |
| 66 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.839 |
0.427 |
| 67 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.833 |
0.382 |
| 68 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
0.826 |
0.160 |
| 69 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.826 |
0.671 |
| 70 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.825 |
0.876 |
| 71 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.824 |
0.658 |
| 72 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.820 |
-0.068 |
| 73 |
266977_at |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
0.812 |
0.575 |
| 74 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.809 |
0.472 |
| 75 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
0.807 |
0.853 |
| 76 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.795 |
0.803 |
| 77 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.794 |
0.770 |
| 78 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.781 |
0.868 |
| 79 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.777 |
0.968 |
| 80 |
257522_at |
AT3G08990
|
[Yippee family putative zinc-binding protein] |
0.772 |
0.156 |
| 81 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
0.769 |
0.298 |
| 82 |
254996_at |
AT4G10390
|
[Protein kinase superfamily protein] |
0.765 |
0.592 |
| 83 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
0.764 |
0.382 |
| 84 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
0.754 |
0.654 |
| 85 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.750 |
1.035 |
| 86 |
254289_at |
AT4G22980
|
unknown |
0.748 |
0.478 |
| 87 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.743 |
0.398 |
| 88 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.738 |
0.649 |
| 89 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.735 |
0.858 |
| 90 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.732 |
0.900 |
| 91 |
247312_at |
AT5G63970
|
RGLG3, RING DOMAIN LIGASE 3 |
0.724 |
0.623 |
| 92 |
252097_at |
AT3G51090
|
unknown |
0.723 |
0.451 |
| 93 |
258209_at |
AT3G14060
|
unknown |
0.722 |
0.801 |
| 94 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.718 |
0.717 |
| 95 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.715 |
-0.003 |
| 96 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.715 |
0.998 |
| 97 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.712 |
1.204 |
| 98 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
0.706 |
0.339 |
| 99 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.704 |
0.765 |
| 100 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.701 |
0.435 |
| 101 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.685 |
0.967 |
| 102 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
0.684 |
-0.181 |
| 103 |
252179_at |
AT3G50760
|
GATL2, galacturonosyltransferase-like 2 |
0.682 |
0.598 |
| 104 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.678 |
0.789 |
| 105 |
263829_at |
AT2G40435
|
unknown |
0.676 |
0.488 |
| 106 |
248490_at |
AT5G50940
|
[RNA-binding KH domain-containing protein] |
0.673 |
0.277 |
| 107 |
252611_at |
AT3G45130
|
LAS1, lanosterol synthase 1 |
0.672 |
0.313 |
| 108 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
0.670 |
0.563 |
| 109 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
0.668 |
0.326 |
| 110 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
0.668 |
0.377 |
| 111 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.664 |
0.239 |
| 112 |
265632_at |
AT2G14290
|
unknown |
0.659 |
0.766 |
| 113 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.659 |
0.511 |
| 114 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.654 |
-0.067 |
| 115 |
259618_at |
AT1G48000
|
AtMYB112, myb domain protein 112, MYB112, myb domain protein 112 |
0.652 |
0.626 |
| 116 |
266456_at |
AT2G22770
|
NAI1 |
0.650 |
0.647 |
| 117 |
249090_at |
AT5G43745
|
unknown |
0.649 |
0.226 |
| 118 |
251063_at |
AT5G01850
|
[Protein kinase superfamily protein] |
0.646 |
0.277 |
| 119 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.643 |
0.260 |
| 120 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.643 |
0.273 |
| 121 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.986 |
-0.767 |
| 122 |
247474_at |
AT5G62280
|
unknown |
-0.971 |
-0.400 |
| 123 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.902 |
-0.736 |
| 124 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.900 |
-0.508 |
| 125 |
250327_at |
AT5G12050
|
unknown |
-0.895 |
-0.252 |
| 126 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.867 |
-0.720 |
| 127 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.830 |
-0.620 |
| 128 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.810 |
-0.050 |
| 129 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.806 |
-0.712 |
| 130 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.804 |
-0.475 |
| 131 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.800 |
-0.035 |
| 132 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.787 |
-0.610 |
| 133 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.771 |
-0.112 |
| 134 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
-0.769 |
0.053 |
| 135 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.764 |
-0.565 |
| 136 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.762 |
-0.214 |
| 137 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.762 |
-0.418 |
| 138 |
247878_at |
AT5G57760
|
unknown |
-0.745 |
-0.372 |
| 139 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.739 |
-0.061 |
| 140 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.735 |
-0.151 |
| 141 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.728 |
-0.278 |
| 142 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.719 |
-0.068 |
| 143 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.719 |
-0.148 |
| 144 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.718 |
-0.295 |
| 145 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.696 |
-0.327 |
| 146 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.695 |
-0.379 |
| 147 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.694 |
-0.829 |
| 148 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.691 |
-0.437 |
| 149 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.685 |
-0.422 |
| 150 |
264238_at |
AT1G54740
|
unknown |
-0.680 |
-0.191 |
| 151 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.675 |
-0.352 |
| 152 |
255779_at |
AT1G18650
|
PDCB3, plasmodesmata callose-binding protein 3 |
-0.671 |
-0.442 |
| 153 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
-0.669 |
0.214 |
| 154 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.659 |
-0.365 |
| 155 |
257204_at |
AT3G23805
|
RALFL24, ralf-like 24 |
-0.653 |
-0.367 |
| 156 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.643 |
-0.270 |
| 157 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.635 |
-0.604 |
| 158 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.633 |
-0.472 |
| 159 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.625 |
-0.296 |
| 160 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.618 |
-0.584 |
| 161 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.615 |
-0.307 |
| 162 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
-0.613 |
-0.351 |
| 163 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.609 |
-0.466 |
| 164 |
261658_at |
AT1G50040
|
unknown |
-0.605 |
-0.112 |
| 165 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.602 |
-0.348 |
| 166 |
261949_at |
AT1G64670
|
BDG1, BODYGUARD1, CED1, 9-CIS EPOXYCAROTENOID DIOXYGENASE DEFECTIVE 1 |
-0.597 |
-0.241 |
| 167 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.596 |
-0.238 |
| 168 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.593 |
-0.713 |
| 169 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.590 |
-0.491 |
| 170 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.587 |
-0.380 |
| 171 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.587 |
0.345 |
| 172 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.587 |
-0.075 |
| 173 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.586 |
-0.033 |
| 174 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.586 |
-0.566 |
| 175 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.584 |
0.128 |
| 176 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.580 |
-0.544 |
| 177 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.577 |
-0.892 |
| 178 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.577 |
-0.414 |
| 179 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.574 |
-0.110 |
| 180 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.573 |
-0.361 |
| 181 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.573 |
-0.240 |
| 182 |
267260_at |
AT2G23130
|
AGP17, arabinogalactan protein 17, ATAGP17 |
-0.573 |
-0.619 |
| 183 |
249755_at |
AT5G24580
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.572 |
-0.697 |
| 184 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.570 |
-0.434 |
| 185 |
247214_at |
AT5G64850
|
unknown |
-0.569 |
-0.044 |
| 186 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.566 |
-0.483 |
| 187 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.565 |
-0.446 |
| 188 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.565 |
0.075 |
| 189 |
259592_at |
AT1G27950
|
LTPG1, glycosylphosphatidylinositol-anchored lipid protein transfer 1 |
-0.560 |
-0.012 |
| 190 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.556 |
-0.463 |
| 191 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.552 |
-0.385 |
| 192 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.551 |
-0.293 |
| 193 |
258468_at |
AT3G06070
|
unknown |
-0.550 |
-0.278 |
| 194 |
250936_at |
AT5G03120
|
unknown |
-0.546 |
-0.598 |
| 195 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.543 |
-0.170 |
| 196 |
263421_at |
AT2G17230
|
EXL5, EXORDIUM like 5 |
-0.541 |
-0.284 |
| 197 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.536 |
-0.295 |
| 198 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.524 |
-0.468 |
| 199 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.523 |
-0.337 |
| 200 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.516 |
-0.422 |
| 201 |
253165_at |
AT4G35320
|
unknown |
-0.516 |
-0.252 |
| 202 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.515 |
-0.190 |
| 203 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.515 |
-0.501 |
| 204 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.513 |
-0.545 |
| 205 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.507 |
-0.289 |
| 206 |
265880_at |
AT2G42300
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.506 |
-0.444 |
| 207 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.503 |
-0.338 |
| 208 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.503 |
-0.689 |
| 209 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.503 |
-0.103 |
| 210 |
245906_at |
AT5G11070
|
unknown |
-0.500 |
-0.016 |
| 211 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.500 |
-0.413 |
| 212 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.497 |
-0.085 |
| 213 |
263194_at |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-0.495 |
-0.045 |
| 214 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
-0.494 |
-0.147 |
| 215 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.493 |
-0.093 |
| 216 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.491 |
-0.401 |
| 217 |
264521_at |
AT1G10020
|
unknown |
-0.491 |
-0.550 |
| 218 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.491 |
-0.342 |
| 219 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.486 |
-0.055 |
| 220 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.485 |
0.180 |
| 221 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.481 |
-0.380 |
| 222 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.478 |
-0.618 |
| 223 |
266921_at |
AT2G45970
|
CYP86A8, cytochrome P450, family 86, subfamily A, polypeptide 8, LCR, LACERATA |
-0.476 |
-0.222 |
| 224 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.473 |
-0.213 |
| 225 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.470 |
-0.396 |
| 226 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
-0.470 |
-0.443 |
| 227 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.467 |
-0.653 |
| 228 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.467 |
-0.160 |
| 229 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.458 |
-0.173 |
| 230 |
257076_at |
AT3G19680
|
unknown |
-0.457 |
-0.221 |
| 231 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.456 |
-0.333 |
| 232 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.455 |
-0.587 |
| 233 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
-0.454 |
-0.245 |
| 234 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.452 |
-0.137 |
| 235 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.451 |
-0.277 |
| 236 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.451 |
-0.431 |
| 237 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.451 |
-0.780 |
| 238 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
-0.451 |
-0.421 |
| 239 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.446 |
-0.477 |