|
probeID |
AGICode |
Annotation |
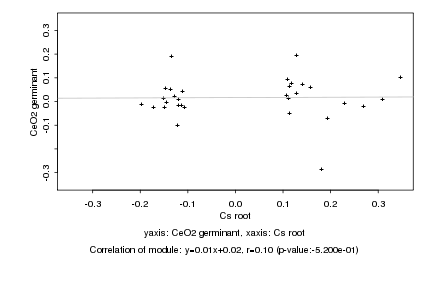
Log2 signal ratio
Cs root |
Log2 signal ratio
CeO2 germinant |
| 1 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.346 |
0.102 |
| 2 |
255138_at |
AT4G08380
|
[Proline-rich extensin-like family protein] |
0.309 |
0.010 |
| 3 |
253352_at |
AT4G33710
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.267 |
-0.021 |
| 4 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.229 |
-0.005 |
| 5 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.192 |
-0.068 |
| 6 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.179 |
-0.285 |
| 7 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
0.156 |
0.061 |
| 8 |
246374_at |
AT1G51840
|
[protein kinase-related] |
0.140 |
0.074 |
| 9 |
255140_x_at |
AT4G08410
|
[Proline-rich extensin-like family protein] |
0.128 |
0.195 |
| 10 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.127 |
0.035 |
| 11 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
0.117 |
0.080 |
| 12 |
253243_at |
AT4G34560
|
unknown |
0.113 |
0.064 |
| 13 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
0.113 |
-0.049 |
| 14 |
258422_at |
AT3G16710
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.111 |
0.013 |
| 15 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
0.109 |
0.097 |
| 16 |
248577_at |
AT5G49870
|
[Mannose-binding lectin superfamily protein] |
0.107 |
0.029 |
| 17 |
257922_at |
AT3G23150
|
ETR2, ethylene response 2 |
-0.199 |
-0.010 |
| 18 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.173 |
-0.022 |
| 19 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
-0.152 |
0.015 |
| 20 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.150 |
-0.024 |
| 21 |
250211_at |
AT5G13880
|
unknown |
-0.147 |
0.055 |
| 22 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
-0.146 |
-0.002 |
| 23 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.137 |
0.053 |
| 24 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.136 |
0.192 |
| 25 |
259485_at |
AT1G15700
|
ATPC2 |
-0.129 |
0.025 |
| 26 |
260668_at |
AT1G19530
|
unknown |
-0.123 |
-0.097 |
| 27 |
262586_at |
AT1G15480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.121 |
0.011 |
| 28 |
261320_at |
AT1G53120
|
[RNA-binding S4 domain-containing protein] |
-0.120 |
-0.013 |
| 29 |
262104_at |
AT1G02910
|
LPA1, LOW PSII ACCUMULATION1 |
-0.114 |
-0.016 |
| 30 |
254350_at |
AT4G22280
|
[F-box/RNI-like superfamily protein] |
-0.113 |
0.046 |
| 31 |
251193_at |
AT3G62910
|
APG3, ALBINO AND PALE GREEN, AtcpRF1, Arabidopsis thaliana chloroplast RF1, cpRF1, chloroplast ribosome release factor 1 |
-0.109 |
-0.025 |