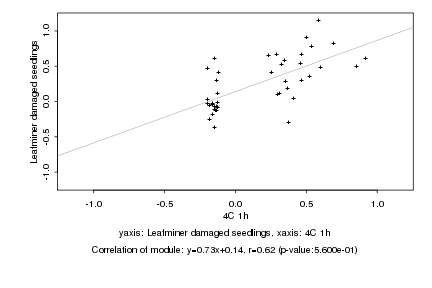
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
4C 1h |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.915 |
0.615 |
| 2 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.850 |
0.507 |
| 3 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.691 |
0.833 |
| 4 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.597 |
0.489 |
| 5 |
263182_at |
AT1G05575
|
unknown |
0.582 |
1.162 |
| 6 |
253643_at |
AT4G29780
|
unknown |
0.535 |
0.791 |
| 7 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.521 |
0.365 |
| 8 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.497 |
0.915 |
| 9 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.466 |
0.310 |
| 10 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
0.462 |
0.667 |
| 11 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.458 |
0.542 |
| 12 |
247406_at |
AT5G62920
|
ARR6, response regulator 6 |
0.406 |
0.044 |
| 13 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
0.373 |
-0.290 |
| 14 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
0.362 |
0.198 |
| 15 |
265184_at |
AT1G23710
|
unknown |
0.352 |
0.294 |
| 16 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.345 |
0.592 |
| 17 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.320 |
0.535 |
| 18 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.310 |
0.115 |
| 19 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.292 |
0.103 |
| 20 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.284 |
0.669 |
| 21 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.249 |
0.417 |
| 22 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.233 |
0.661 |
| 23 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.199 |
0.469 |
| 24 |
249206_at |
AT5G42630
|
ATS, ABERRANT TESTA SHAPE, KAN4, KANADI 4 |
-0.198 |
-0.014 |
| 25 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.198 |
0.042 |
| 26 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.190 |
-0.047 |
| 27 |
267148_at |
AT2G23470
|
RUS4, ROOT UV-B SENSITIVE 4 |
-0.189 |
-0.253 |
| 28 |
255873_at |
AT2G30340
|
LBD13, LOB domain-containing protein 13 |
-0.167 |
-0.029 |
| 29 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.164 |
-0.015 |
| 30 |
262150_at |
AT1G52520
|
FRS6, FAR1-related sequence 6 |
-0.162 |
-0.178 |
| 31 |
253786_at |
AT4G28650
|
[Leucine-rich repeat transmembrane protein kinase family protein] |
-0.154 |
-0.105 |
| 32 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.154 |
0.622 |
| 33 |
248051_at |
AT5G56110
|
AtMYB103, myb domain protein 103, ATMYB80, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 80, MS188, MALE STERILE 188, MYB103, myb domain protein 103, MYB80 |
-0.154 |
-0.058 |
| 34 |
253243_at |
AT4G34560
|
unknown |
-0.149 |
-0.359 |
| 35 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.142 |
-0.119 |
| 36 |
254194_at |
AT4G23980
|
ARF9, auxin response factor 9 |
-0.137 |
-0.124 |
| 37 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
-0.135 |
0.300 |
| 38 |
263869_at |
AT2G22000
|
PROPEP6, elicitor peptide 6 precursor |
-0.134 |
-0.063 |
| 39 |
250572_at |
AT5G08210
|
MIR834A, microRNA834A |
-0.133 |
-0.072 |
| 40 |
262664_at |
AT1G13970
|
unknown |
-0.132 |
-0.002 |
| 41 |
256667_at |
AT3G32180
|
unknown |
-0.128 |
-0.072 |
| 42 |
261839_at |
AT1G16040
|
unknown |
-0.127 |
0.117 |
| 43 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
-0.122 |
0.425 |