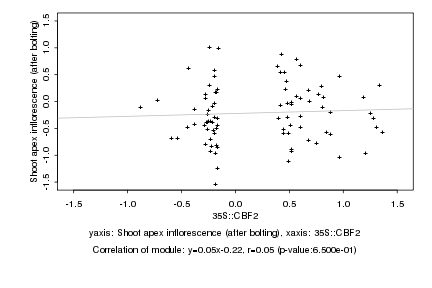
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::CBF2 |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.355 |
-0.573 |
| 2 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
1.332 |
0.303 |
| 3 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
1.302 |
-0.476 |
| 4 |
256114_at |
AT1G16850
|
unknown |
1.273 |
-0.312 |
| 5 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
1.245 |
-0.222 |
| 6 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
1.204 |
-0.964 |
| 7 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
1.179 |
0.082 |
| 8 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.962 |
0.480 |
| 9 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.961 |
-1.041 |
| 10 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.881 |
-0.199 |
| 11 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.876 |
-0.606 |
| 12 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.837 |
-0.567 |
| 13 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.811 |
0.091 |
| 14 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.807 |
-0.111 |
| 15 |
251753_at |
AT3G55760
|
unknown |
0.793 |
0.291 |
| 16 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
0.766 |
0.139 |
| 17 |
260264_at |
AT1G68500
|
unknown |
0.751 |
-0.773 |
| 18 |
261048_at |
AT1G01420
|
UGT72B3, UDP-glucosyl transferase 72B3 |
0.681 |
0.016 |
| 19 |
250467_at |
AT5G10100
|
TPPI, trehalose-6-phosphate phosphatase I |
0.673 |
-0.709 |
| 20 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.669 |
0.221 |
| 21 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.599 |
0.679 |
| 22 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.599 |
-0.267 |
| 23 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.598 |
0.067 |
| 24 |
245533_at |
AT4G15130
|
ATCCT2, CCT2, phosphorylcholine cytidylyltransferase2 |
0.597 |
-0.475 |
| 25 |
247399_at |
AT5G62960
|
unknown |
0.565 |
0.094 |
| 26 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
0.560 |
0.785 |
| 27 |
263352_at |
AT2G22080
|
unknown |
0.518 |
-0.004 |
| 28 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.518 |
-0.926 |
| 29 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.511 |
-0.891 |
| 30 |
258078_at |
AT3G25870
|
unknown |
0.511 |
-0.049 |
| 31 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.508 |
-0.428 |
| 32 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.488 |
-1.105 |
| 33 |
267538_at |
AT2G41870
|
[Remorin family protein] |
0.487 |
-0.580 |
| 34 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.479 |
-0.288 |
| 35 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.476 |
-0.028 |
| 36 |
264989_at |
AT1G27200
|
unknown |
0.470 |
0.378 |
| 37 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.461 |
0.230 |
| 38 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.451 |
0.546 |
| 39 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.443 |
-0.585 |
| 40 |
259426_at |
AT1G01470
|
LEA14, LATE EMBRYOGENESIS ABUNDANT 14, LSR3, LIGHT STRESS-REGULATED 3 |
0.439 |
-0.505 |
| 41 |
252127_at |
AT3G50960
|
PLP3a, phosducin-like protein 3 homolog |
0.427 |
0.882 |
| 42 |
266259_at |
AT2G27830
|
unknown |
0.416 |
-0.063 |
| 43 |
254850_at |
AT4G12000
|
[SNARE associated Golgi protein family] |
0.416 |
0.544 |
| 44 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
0.396 |
-0.303 |
| 45 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.384 |
0.660 |
| 46 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.885 |
-0.110 |
| 47 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.730 |
0.033 |
| 48 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.597 |
-0.686 |
| 49 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.540 |
-0.684 |
| 50 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.453 |
-0.474 |
| 51 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.440 |
0.632 |
| 52 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.382 |
-0.415 |
| 53 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.381 |
-0.131 |
| 54 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.291 |
-0.437 |
| 55 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.285 |
0.144 |
| 56 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.285 |
0.068 |
| 57 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.284 |
-0.787 |
| 58 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.269 |
-0.373 |
| 59 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.262 |
-0.237 |
| 60 |
247683_at |
AT5G59660
|
[Leucine-rich repeat protein kinase family protein] |
-0.262 |
-0.380 |
| 61 |
261247_at |
AT1G20070
|
unknown |
-0.261 |
-0.503 |
| 62 |
252951_at |
AT4G38700
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.252 |
-0.157 |
| 63 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.250 |
-0.359 |
| 64 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.248 |
0.301 |
| 65 |
251142_at |
AT5G01015
|
unknown |
-0.247 |
1.017 |
| 66 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.236 |
-0.925 |
| 67 |
266275_at |
AT2G29370
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.234 |
-0.695 |
| 68 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
-0.233 |
-0.367 |
| 69 |
253305_at |
AT4G33666
|
unknown |
-0.225 |
-0.821 |
| 70 |
249726_at |
AT5G35480
|
unknown |
-0.222 |
-0.091 |
| 71 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.217 |
-0.385 |
| 72 |
263470_at |
AT2G31900
|
ATMYO5, MYOSIN 5, ATXIF, MYOSIN XI F, XIF, myosin-like protein XIF |
-0.208 |
-0.533 |
| 73 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.203 |
0.473 |
| 74 |
252906_at |
AT4G39640
|
GGT1, gamma-glutamyl transpeptidase 1 |
-0.203 |
0.596 |
| 75 |
246552_at |
AT5G15420
|
unknown |
-0.200 |
-0.285 |
| 76 |
255028_at |
AT4G09890
|
unknown |
-0.198 |
-0.593 |
| 77 |
258727_at |
AT3G11930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.195 |
-0.036 |
| 78 |
256843_at |
AT3G31940
|
unknown |
-0.191 |
-1.527 |
| 79 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.188 |
0.179 |
| 80 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.185 |
-0.955 |
| 81 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.181 |
-0.502 |
| 82 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.178 |
-0.805 |
| 83 |
247633_at |
AT5G60470
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.177 |
0.183 |
| 84 |
259572_at |
AT1G20400
|
unknown |
-0.173 |
-0.844 |
| 85 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-0.172 |
-1.247 |
| 86 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
-0.172 |
0.224 |
| 87 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.171 |
-0.298 |
| 88 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
-0.170 |
-0.434 |
| 89 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
-0.164 |
1.005 |