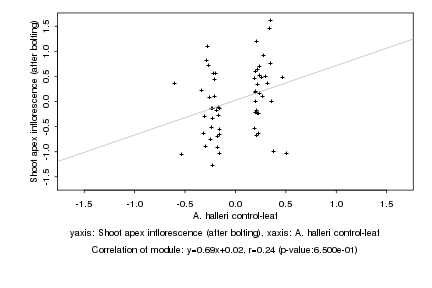
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
A. halleri control-leaf |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.506 |
-1.027 |
| 2 |
263978_at |
AT2G42680
|
ATMBF1A, ARABIDOPSIS THALIANA MULTIPROTEIN BRIDGING FACTOR 1A, MBF1A, multiprotein bridging factor 1A |
0.462 |
0.490 |
| 3 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.371 |
-0.995 |
| 4 |
247653_at |
AT5G59950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.351 |
0.002 |
| 5 |
246730_at |
AT5G28060
|
[Ribosomal protein S24e family protein] |
0.343 |
1.633 |
| 6 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.342 |
0.773 |
| 7 |
253273_at |
AT4G34180
|
[Cyclase family protein] |
0.332 |
1.462 |
| 8 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.317 |
0.371 |
| 9 |
258455_at |
AT3G22440
|
[FRIGIDA-like protein] |
0.295 |
0.514 |
| 10 |
252565_at |
AT3G46000
|
ADF2, actin depolymerizing factor 2 |
0.269 |
0.927 |
| 11 |
261561_at |
AT1G01730
|
unknown |
0.262 |
0.109 |
| 12 |
258173_at |
AT3G21630
|
AtCERK1, AtLYK1, CERK1, chitin elicitor receptor kinase 1, LYK1, LysM-containing receptor-like kinase 1, LYSM RLK1, LYSM DOMAIN RECEPTOR-LIKE KINASE 1 |
0.256 |
0.488 |
| 13 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
0.234 |
0.163 |
| 14 |
249861_at |
AT5G22875
|
unknown |
0.233 |
0.538 |
| 15 |
246770_at |
AT5G27460
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.231 |
0.713 |
| 16 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
0.220 |
-0.235 |
| 17 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.219 |
-0.636 |
| 18 |
265375_at |
AT2G06530
|
VPS2.1 |
0.214 |
-0.227 |
| 19 |
256240_at |
AT3G12600
|
atnudt16, nudix hydrolase homolog 16, NUDT16, nudix hydrolase homolog 16 |
0.210 |
0.642 |
| 20 |
256058_at |
AT1G07240
|
UGT71C5, UDP-glucosyl transferase 71C5 |
0.210 |
0.351 |
| 21 |
253404_at |
AT4G32840
|
PFK6, phosphofructokinase 6 |
0.206 |
1.204 |
| 22 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.205 |
-0.172 |
| 23 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.202 |
-0.671 |
| 24 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.195 |
-0.211 |
| 25 |
259230_at |
AT3G07780
|
OBE1, OBERON1 |
0.194 |
0.014 |
| 26 |
246796_at |
AT5G26770
|
unknown |
0.192 |
0.195 |
| 27 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.191 |
0.219 |
| 28 |
256899_at |
AT3G24660
|
TMKL1, transmembrane kinase-like 1 |
0.190 |
0.604 |
| 29 |
267486_at |
AT2G02800
|
APK2B, protein kinase 2B, Kin2, kinase 2 |
0.186 |
0.476 |
| 30 |
247124_at |
AT5G66060
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.185 |
-0.528 |
| 31 |
246463_at |
AT5G16970
|
AER, alkenal reductase, AT-AER, alkenal reductase |
-0.605 |
0.369 |
| 32 |
255273_at |
AT4G05280
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G27780.1)] |
-0.535 |
-1.044 |
| 33 |
247227_at |
AT5G65130
|
[Integrase-type DNA-binding superfamily protein] |
-0.338 |
0.235 |
| 34 |
251401_at |
AT3G60270
|
[Cupredoxin superfamily protein] |
-0.320 |
-0.622 |
| 35 |
249432_at |
AT5G39930
|
CLPS5, CLP1-similar protein 5 |
-0.307 |
-0.298 |
| 36 |
254841_at |
AT4G11940
|
unknown |
-0.297 |
-0.894 |
| 37 |
257544_at |
AT3G20880
|
WIP4, WIP domain protein 4 |
-0.293 |
0.829 |
| 38 |
250711_at |
AT5G06110
|
[DnaJ domain ] |
-0.279 |
1.106 |
| 39 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.269 |
0.723 |
| 40 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.263 |
0.085 |
| 41 |
261736_at |
AT1G47810
|
[F-box and associated interaction domains-containing protein] |
-0.255 |
-0.750 |
| 42 |
265153_at |
AT1G30950
|
UFO, UNUSUAL FLORAL ORGANS |
-0.240 |
-0.507 |
| 43 |
247349_at |
AT5G63820
|
unknown |
-0.238 |
-0.130 |
| 44 |
246168_at |
AT5G32460
|
[Transcriptional factor B3 family protein] |
-0.237 |
-1.263 |
| 45 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.233 |
-0.129 |
| 46 |
245614_at |
AT4G14460
|
[copia-like retrotransposon family, has a 4.2e-253 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.230 |
-0.337 |
| 47 |
247613_at |
AT5G60740
|
ABCG28, ATP-binding cassette G28 |
-0.219 |
0.571 |
| 48 |
255004_at |
AT4G09970
|
unknown |
-0.211 |
0.450 |
| 49 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
-0.211 |
0.112 |
| 50 |
266015_at |
AT2G24190
|
SDR2, short-chain dehydrogenase/reductase 2 |
-0.200 |
0.567 |
| 51 |
251788_at |
AT3G55420
|
unknown |
-0.197 |
-0.163 |
| 52 |
263038_at |
AT1G23270
|
unknown |
-0.178 |
-0.901 |
| 53 |
252839_at |
AT3G42100
|
[similar to AT hook motif-containing protein-related [Arabidopsis thaliana] (TAIR:AT1G35940.1)] |
-0.178 |
-0.678 |
| 54 |
246618_at |
AT5G36280
|
unknown |
-0.174 |
-0.276 |
| 55 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.172 |
-0.106 |
| 56 |
259881_at |
AT1G76820
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
-0.168 |
-0.549 |
| 57 |
256834_at |
AT3G22920
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.166 |
-1.031 |
| 58 |
255287_at |
AT4G04660
|
[pseudogene] |
-0.166 |
-0.637 |
| 59 |
250481_at |
AT5G10310
|
unknown |
-0.164 |
-0.122 |