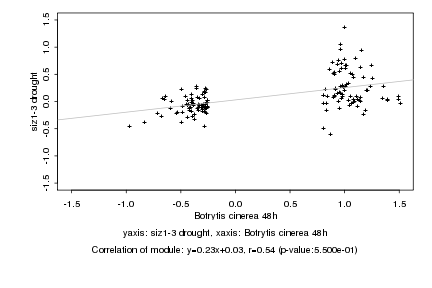
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Botrytis cinerea 48h |
Log2 signal ratio
siz1-3 drought |
| 1 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
1.508 |
-0.033 |
| 2 |
252487_at |
AT3G46660
|
UGT76E12, UDP-glucosyl transferase 76E12 |
1.488 |
0.094 |
| 3 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.487 |
0.042 |
| 4 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.389 |
0.053 |
| 5 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.388 |
0.025 |
| 6 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.355 |
0.287 |
| 7 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
1.340 |
0.063 |
| 8 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
1.247 |
0.424 |
| 9 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
1.237 |
0.668 |
| 10 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
1.230 |
0.285 |
| 11 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
1.207 |
0.221 |
| 12 |
251293_at |
AT3G61930
|
unknown |
1.194 |
0.211 |
| 13 |
262640_at |
AT1G62760
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
1.182 |
-0.150 |
| 14 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
1.172 |
0.444 |
| 15 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
1.169 |
-0.221 |
| 16 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
1.149 |
0.953 |
| 17 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.145 |
0.637 |
| 18 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.143 |
0.010 |
| 19 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
1.139 |
0.079 |
| 20 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
1.129 |
0.022 |
| 21 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
1.113 |
-0.081 |
| 22 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
1.112 |
0.049 |
| 23 |
259150_at |
AT3G10320
|
[Glycosyltransferase family 61 protein] |
1.100 |
0.104 |
| 24 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
1.091 |
0.793 |
| 25 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
1.089 |
0.005 |
| 26 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.078 |
0.458 |
| 27 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
1.078 |
-0.002 |
| 28 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
1.075 |
0.044 |
| 29 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
1.066 |
0.509 |
| 30 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
1.061 |
-0.026 |
| 31 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
1.053 |
0.531 |
| 32 |
245076_at |
AT2G23170
|
GH3.3 |
1.046 |
0.095 |
| 33 |
258182_at |
AT3G21500
|
DXL1, DXS-like 1, DXPS1, 1-deoxy-D-xylulose 5-phosphate synthase 1, DXS2, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase 2 |
1.041 |
-0.059 |
| 34 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.027 |
0.332 |
| 35 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
1.027 |
0.025 |
| 36 |
251456_at |
AT3G60120
|
BGLU27, beta glucosidase 27 |
1.014 |
0.667 |
| 37 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
1.013 |
0.318 |
| 38 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
1.006 |
0.616 |
| 39 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
1.000 |
0.667 |
| 40 |
246984_at |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
1.000 |
0.290 |
| 41 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
0.997 |
0.215 |
| 42 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.993 |
0.790 |
| 43 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.990 |
1.366 |
| 44 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.986 |
0.266 |
| 45 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.979 |
0.299 |
| 46 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.974 |
0.095 |
| 47 |
264580_at |
AT1G05340
|
unknown |
0.973 |
0.147 |
| 48 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.971 |
0.621 |
| 49 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.966 |
0.067 |
| 50 |
253060_at |
AT4G37710
|
[VQ motif-containing protein] |
0.965 |
0.714 |
| 51 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
0.960 |
0.968 |
| 52 |
256024_at |
AT1G58340
|
BCD1, BUSH-AND-CHLOROTIC-DWARF 1, ZF14, ZRZ, ZRIZI |
0.960 |
0.160 |
| 53 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.959 |
1.050 |
| 54 |
260406_at |
AT1G69920
|
ATGSTU12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 12, GSTU12, glutathione S-transferase TAU 12 |
0.955 |
0.148 |
| 55 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.954 |
0.281 |
| 56 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
0.952 |
-0.126 |
| 57 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.947 |
0.561 |
| 58 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.946 |
0.167 |
| 59 |
249454_at |
AT5G39520
|
unknown |
0.940 |
0.006 |
| 60 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.939 |
0.768 |
| 61 |
253505_at |
AT4G31970
|
CYP82C2, cytochrome P450, family 82, subfamily C, polypeptide 2 |
0.928 |
0.161 |
| 62 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.927 |
0.685 |
| 63 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.912 |
0.230 |
| 64 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
0.910 |
0.253 |
| 65 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.906 |
0.108 |
| 66 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.904 |
0.536 |
| 67 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.903 |
0.509 |
| 68 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.900 |
0.115 |
| 69 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.890 |
0.085 |
| 70 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.890 |
0.528 |
| 71 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.888 |
0.720 |
| 72 |
263829_at |
AT2G40435
|
unknown |
0.862 |
-0.590 |
| 73 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.859 |
0.597 |
| 74 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.834 |
0.094 |
| 75 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.830 |
-0.162 |
| 76 |
247799_at |
AT5G58840
|
[Subtilase family protein] |
0.826 |
-0.032 |
| 77 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.821 |
0.236 |
| 78 |
245444_at |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
0.805 |
-0.484 |
| 79 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.799 |
0.117 |
| 80 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.799 |
-0.030 |
| 81 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.798 |
0.112 |
| 82 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.971 |
-0.457 |
| 83 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.834 |
-0.374 |
| 84 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.714 |
-0.211 |
| 85 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.683 |
-0.258 |
| 86 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.677 |
0.068 |
| 87 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.657 |
0.053 |
| 88 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.641 |
0.093 |
| 89 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.602 |
-0.115 |
| 90 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.591 |
0.002 |
| 91 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.545 |
-0.213 |
| 92 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.535 |
-0.197 |
| 93 |
264238_at |
AT1G54740
|
unknown |
-0.503 |
0.230 |
| 94 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.501 |
-0.380 |
| 95 |
249932_at |
AT5G22390
|
unknown |
-0.489 |
-0.190 |
| 96 |
251142_at |
AT5G01015
|
unknown |
-0.488 |
-0.085 |
| 97 |
264605_at |
AT1G04550
|
BDL, BODENLOS, IAA12, indole-3-acetic acid inducible 12 |
-0.465 |
0.094 |
| 98 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.455 |
-0.053 |
| 99 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.446 |
0.024 |
| 100 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.443 |
-0.283 |
| 101 |
265387_at |
AT2G20670
|
unknown |
-0.443 |
-0.066 |
| 102 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.435 |
-0.051 |
| 103 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
-0.425 |
-0.163 |
| 104 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.424 |
-0.122 |
| 105 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.422 |
-0.115 |
| 106 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
-0.419 |
-0.106 |
| 107 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
-0.418 |
-0.102 |
| 108 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.409 |
0.140 |
| 109 |
249392_at |
AT5G40150
|
[Peroxidase superfamily protein] |
-0.407 |
-0.182 |
| 110 |
253165_at |
AT4G35320
|
unknown |
-0.406 |
-0.013 |
| 111 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.404 |
0.069 |
| 112 |
245318_at |
AT4G16980
|
[arabinogalactan-protein family] |
-0.400 |
-0.048 |
| 113 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.396 |
0.023 |
| 114 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
-0.395 |
-0.274 |
| 115 |
267339_at |
AT2G39870
|
unknown |
-0.389 |
-0.069 |
| 116 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.385 |
-0.035 |
| 117 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.380 |
-0.235 |
| 118 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.375 |
-0.317 |
| 119 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.359 |
0.249 |
| 120 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.359 |
0.278 |
| 121 |
253062_at |
AT4G37590
|
MEL1, MAB4/ENP/NPY1-LIKE 1, NPY5, NAKED PINS IN YUC MUTANTS 5 |
-0.354 |
-0.126 |
| 122 |
262421_at |
AT1G50290
|
unknown |
-0.353 |
0.077 |
| 123 |
254145_at |
AT4G24700
|
unknown |
-0.346 |
-0.107 |
| 124 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.343 |
-0.045 |
| 125 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
-0.342 |
-0.155 |
| 126 |
261252_at |
AT1G05810
|
ARA, ARA-1, ATRAB11D, ATRABA5E, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG A5E, RABA5E, RAB GTPase homolog A5E |
-0.338 |
-0.063 |
| 127 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.335 |
0.066 |
| 128 |
256396_at |
AT3G06150
|
unknown |
-0.333 |
-0.055 |
| 129 |
250972_at |
AT5G02840
|
LCL1, LHY/CCA1-like 1 |
-0.318 |
-0.059 |
| 130 |
254810_at |
AT4G12390
|
PME1, pectin methylesterase inhibitor 1 |
-0.312 |
-0.057 |
| 131 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.311 |
-0.094 |
| 132 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
-0.307 |
-0.107 |
| 133 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.305 |
-0.055 |
| 134 |
258535_at |
AT3G06750
|
[hydroxyproline-rich glycoprotein family protein] |
-0.305 |
-0.166 |
| 135 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.304 |
-0.144 |
| 136 |
251946_at |
AT3G53540
|
TRM19, TON1 Recruiting Motif 19 |
-0.303 |
0.137 |
| 137 |
259958_at |
AT1G53730
|
SRF6, STRUBBELIG-receptor family 6 |
-0.294 |
-0.084 |
| 138 |
259358_at |
AT1G13250
|
GATL3, galacturonosyltransferase-like 3 |
-0.293 |
-0.039 |
| 139 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.291 |
0.187 |
| 140 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.291 |
-0.457 |
| 141 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
-0.290 |
-0.104 |
| 142 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.290 |
0.087 |
| 143 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
-0.290 |
0.170 |
| 144 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.289 |
-0.168 |
| 145 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.282 |
-0.040 |
| 146 |
264379_at |
AT2G25200
|
unknown |
-0.280 |
-0.190 |
| 147 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
-0.279 |
-0.092 |
| 148 |
259523_at |
AT1G12500
|
[Nucleotide-sugar transporter family protein] |
-0.279 |
-0.126 |
| 149 |
260696_at |
AT1G32520
|
unknown |
-0.279 |
0.170 |
| 150 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.275 |
0.250 |
| 151 |
259858_at |
AT1G68400
|
[leucine-rich repeat transmembrane protein kinase family protein] |
-0.275 |
-0.044 |
| 152 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.273 |
0.235 |
| 153 |
254448_at |
AT4G20870
|
ATFAH2, ARABIDOPSIS FATTY ACID HYDROXYLASE 2, FAH2, fatty acid hydroxylase 2 |
-0.272 |
-0.004 |
| 154 |
264573_at |
AT1G05310
|
[Pectin lyase-like superfamily protein] |
-0.271 |
-0.220 |
| 155 |
254787_at |
AT4G12690
|
unknown |
-0.266 |
-0.114 |
| 156 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.265 |
0.034 |
| 157 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.265 |
-0.005 |
| 158 |
256262_at |
AT3G12150
|
unknown |
-0.264 |
-0.100 |
| 159 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
-0.262 |
-0.099 |
| 160 |
250696_at |
AT5G06790
|
unknown |
-0.262 |
0.024 |
| 161 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.262 |
-0.082 |