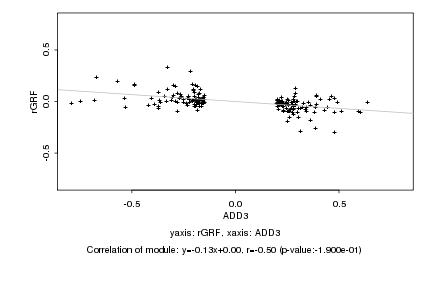
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ADD3 |
Log2 signal ratio
rGRF |
| 1 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
0.634 |
-0.009 |
| 2 |
266078_at |
AT2G40670
|
ARR16, response regulator 16, RR16, response regulator 16 |
0.603 |
-0.106 |
| 3 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
0.591 |
-0.092 |
| 4 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.510 |
-0.094 |
| 5 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
0.488 |
-0.000 |
| 6 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.478 |
-0.102 |
| 7 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.475 |
-0.298 |
| 8 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.475 |
0.036 |
| 9 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.461 |
0.053 |
| 10 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
0.450 |
0.022 |
| 11 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
0.441 |
-0.054 |
| 12 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
0.429 |
-0.085 |
| 13 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.410 |
0.020 |
| 14 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.391 |
0.066 |
| 15 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.388 |
-0.028 |
| 16 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.387 |
0.051 |
| 17 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.386 |
-0.254 |
| 18 |
261653_at |
AT1G01900
|
ATSBT1.1, SBTI1.1 |
0.386 |
-0.056 |
| 19 |
265539_at |
AT2G15830
|
unknown |
0.381 |
-0.100 |
| 20 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.362 |
-0.179 |
| 21 |
248201_at |
AT5G54180
|
PTAC15, plastid transcriptionally active 15 |
0.360 |
-0.038 |
| 22 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
0.351 |
-0.001 |
| 23 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
0.343 |
-0.090 |
| 24 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
0.341 |
-0.054 |
| 25 |
262693_at |
AT1G62780
|
unknown |
0.337 |
-0.072 |
| 26 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
0.324 |
-0.019 |
| 27 |
258530_at |
AT3G06840
|
unknown |
0.320 |
-0.051 |
| 28 |
257909_at |
AT3G25480
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.311 |
-0.067 |
| 29 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.311 |
-0.285 |
| 30 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
0.304 |
-0.059 |
| 31 |
247486_at |
AT5G62140
|
unknown |
0.303 |
-0.153 |
| 32 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.299 |
-0.103 |
| 33 |
262796_at |
AT1G20850
|
XCP2, xylem cysteine peptidase 2 |
0.298 |
0.002 |
| 34 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
0.292 |
0.004 |
| 35 |
252278_at |
AT3G49530
|
ANAC062, NAC domain containing protein 62, NAC062, NAC domain containing protein 62, NTL6, NTM1 (NAC WITH TRANSMEMBRANE MOTIF 1)-LIKE 6 |
0.289 |
-0.028 |
| 36 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
0.289 |
-0.031 |
| 37 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
0.288 |
0.135 |
| 38 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.287 |
0.084 |
| 39 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.287 |
-0.030 |
| 40 |
262236_at |
AT1G48330
|
unknown |
0.285 |
0.056 |
| 41 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
0.284 |
0.009 |
| 42 |
258222_at |
AT3G15680
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
0.280 |
-0.040 |
| 43 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.279 |
-0.064 |
| 44 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.278 |
0.020 |
| 45 |
255647_at |
AT4G00900
|
ATECA2, ARABIDOPSIS THALIANA ER-TYPE CA2+-ATPASE 2, ECA2, ER-type Ca2+-ATPase 2 |
0.277 |
-0.066 |
| 46 |
264343_at |
AT1G11850
|
unknown |
0.276 |
-0.076 |
| 47 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.276 |
0.009 |
| 48 |
264636_at |
AT1G65490
|
unknown |
0.276 |
-0.118 |
| 49 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
0.275 |
-0.028 |
| 50 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
0.273 |
-0.104 |
| 51 |
248908_at |
AT5G45800
|
MEE62, maternal effect embryo arrest 62 |
0.269 |
-0.002 |
| 52 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.268 |
-0.065 |
| 53 |
245027_at |
AT2G26550
|
HO2, heme oxygenase 2 |
0.267 |
-0.066 |
| 54 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
0.264 |
-0.074 |
| 55 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
0.259 |
-0.091 |
| 56 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.257 |
-0.153 |
| 57 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.254 |
-0.031 |
| 58 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.253 |
-0.020 |
| 59 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.252 |
-0.096 |
| 60 |
266221_at |
AT2G28760
|
UXS6, UDP-XYL synthase 6 |
0.252 |
0.010 |
| 61 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
0.251 |
-0.096 |
| 62 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
0.250 |
0.027 |
| 63 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
0.247 |
-0.185 |
| 64 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
0.245 |
-0.000 |
| 65 |
250146_at |
AT5G14660
|
ATDEF2, DEF2, PDF1B, peptide deformylase 1B |
0.245 |
-0.060 |
| 66 |
252215_at |
AT3G50240
|
KICP-02 |
0.243 |
-0.016 |
| 67 |
251050_at |
AT5G02440
|
unknown |
0.241 |
-0.012 |
| 68 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
0.239 |
-0.020 |
| 69 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
0.232 |
-0.043 |
| 70 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.232 |
-0.092 |
| 71 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.231 |
-0.013 |
| 72 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
0.230 |
-0.022 |
| 73 |
253589_at |
AT4G30825
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.229 |
-0.082 |
| 74 |
251631_at |
AT3G57430
|
OTP84, ORGANELLE TRANSCRIPT PROCESSING 84 |
0.228 |
-0.088 |
| 75 |
265484_at |
AT2G15820
|
OTP51, ORGANELLE TRANSCRIPT PROCESSING 51 |
0.227 |
-0.048 |
| 76 |
258183_at |
AT3G21550
|
AtDMP2, Arabidopsis thaliana DUF679 domain membrane protein 2, DMP2, DUF679 domain membrane protein 2 |
0.223 |
0.007 |
| 77 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.221 |
0.048 |
| 78 |
252001_at |
AT3G52750
|
FTSZ2-2 |
0.220 |
-0.017 |
| 79 |
260325_at |
AT1G63940
|
MDAR6, monodehydroascorbate reductase 6 |
0.220 |
0.009 |
| 80 |
261602_at |
AT1G49630
|
ATPREP2, presequence protease 2, PREP2, presequence protease 2 |
0.218 |
-0.028 |
| 81 |
246265_at |
AT1G31860
|
AT-IE, HISN2, HISTIDINE BIOSYNTHESIS 2 |
0.213 |
-0.021 |
| 82 |
253297_at |
AT4G33810
|
[Glycosyl hydrolase superfamily protein] |
0.212 |
-0.008 |
| 83 |
251151_at |
AT3G63170
|
AtFAP1, FAP1, fatty-acid-binding protein 1 |
0.210 |
0.012 |
| 84 |
248380_at |
AT5G51820
|
ATPGMP, ARABIDOPSIS THALIANA PHOSPHOGLUCOMUTASE, PGM, phosphoglucomutase, PGM1, STF1, STARCH-FREE 1 |
0.210 |
-0.024 |
| 85 |
251753_at |
AT3G55760
|
unknown |
0.209 |
-0.029 |
| 86 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.208 |
0.005 |
| 87 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
0.206 |
-0.019 |
| 88 |
248023_at |
AT5G56450
|
PM-ANT |
0.206 |
-0.025 |
| 89 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
0.205 |
-0.077 |
| 90 |
253317_at |
AT4G33960
|
unknown |
0.205 |
-0.013 |
| 91 |
260065_at |
AT1G73760
|
[RING/U-box superfamily protein] |
0.204 |
-0.013 |
| 92 |
262081_at |
AT1G59540
|
ZCF125 |
0.203 |
0.019 |
| 93 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
0.203 |
-0.040 |
| 94 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
0.203 |
0.012 |
| 95 |
248057_at |
AT5G55520
|
unknown |
0.203 |
0.000 |
| 96 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
0.199 |
0.028 |
| 97 |
247586_at |
AT5G60660
|
PIP2;4, plasma membrane intrinsic protein 2;4, PIP2F |
0.198 |
-0.018 |
| 98 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
-0.795 |
-0.019 |
| 99 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
-0.752 |
0.005 |
| 100 |
248260_at |
AT5G53240
|
unknown |
-0.681 |
0.012 |
| 101 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.672 |
0.237 |
| 102 |
265387_at |
AT2G20670
|
unknown |
-0.574 |
0.201 |
| 103 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.540 |
0.036 |
| 104 |
247515_at |
AT5G61740
|
ABCA10, ATP-binding cassette A10, ATATH14, ARABIDOPSIS THALIANA ABC2 HOMOLOG 14, ATH14, ABC2 homolog 14 |
-0.533 |
-0.051 |
| 105 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.491 |
0.167 |
| 106 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.491 |
0.164 |
| 107 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.422 |
-0.033 |
| 108 |
254287_at |
AT4G22960
|
unknown |
-0.407 |
0.031 |
| 109 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
-0.395 |
-0.021 |
| 110 |
246103_at |
AT5G28640
|
AN3, ANGUSTIFOLIA 3, ATGIF1, ARABIDOPSIS GRF1-INTERACTING FACTOR 1, GIF, GRF1-INTERACTING FACTOR, GIF1, GRF1-INTERACTING FACTOR 1 |
-0.376 |
-0.041 |
| 111 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.375 |
0.092 |
| 112 |
256188_at |
AT1G30160
|
unknown |
-0.373 |
-0.063 |
| 113 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
-0.370 |
0.010 |
| 114 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.364 |
-0.005 |
| 115 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
-0.344 |
0.058 |
| 116 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-0.333 |
0.001 |
| 117 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.330 |
0.124 |
| 118 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
-0.329 |
0.335 |
| 119 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.313 |
0.015 |
| 120 |
263633_at |
AT2G04890
|
SCL21, SCARECROW-like 21 |
-0.306 |
0.040 |
| 121 |
266456_at |
AT2G22770
|
NAI1 |
-0.303 |
0.164 |
| 122 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
-0.302 |
0.061 |
| 123 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.294 |
0.151 |
| 124 |
265988_at |
AT2G24255
|
unknown |
-0.291 |
0.002 |
| 125 |
250380_at |
AT5G11600
|
unknown |
-0.284 |
0.083 |
| 126 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
-0.284 |
-0.003 |
| 127 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.280 |
-0.093 |
| 128 |
257381_at |
AT2G37950
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.272 |
0.032 |
| 129 |
258263_at |
AT3G15780
|
unknown |
-0.268 |
0.045 |
| 130 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.267 |
0.070 |
| 131 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
-0.265 |
0.053 |
| 132 |
266797_at |
AT2G22840
|
AtGRF1, growth-regulating factor 1, GRF1, growth-regulating factor 1 |
-0.255 |
0.028 |
| 133 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
-0.255 |
-0.009 |
| 134 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.238 |
-0.017 |
| 135 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.236 |
-0.030 |
| 136 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.232 |
0.038 |
| 137 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.231 |
0.052 |
| 138 |
257074_at |
AT3G19660
|
unknown |
-0.229 |
0.052 |
| 139 |
247317_at |
AT5G64060
|
anac103, NAC domain containing protein 103, NAC103, NAC domain containing protein 103 |
-0.224 |
-0.003 |
| 140 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.222 |
0.022 |
| 141 |
263914_at |
AT2G36400
|
AtGRF3, growth-regulating factor 3, GRF3, growth-regulating factor 3 |
-0.220 |
0.298 |
| 142 |
247835_at |
AT5G57910
|
unknown |
-0.212 |
0.169 |
| 143 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.210 |
0.015 |
| 144 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
-0.209 |
0.005 |
| 145 |
256597_at |
AT3G28500
|
[60S acidic ribosomal protein family] |
-0.208 |
0.004 |
| 146 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.206 |
0.116 |
| 147 |
264379_at |
AT2G25200
|
unknown |
-0.204 |
0.123 |
| 148 |
266360_at |
AT2G32250
|
FRS2, FAR1-related sequence 2 |
-0.203 |
0.014 |
| 149 |
258604_at |
AT3G02980
|
MCC1, MEIOTIC CONTROL OF CROSSOVERS1 |
-0.202 |
-0.043 |
| 150 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.200 |
0.052 |
| 151 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
-0.200 |
0.090 |
| 152 |
261145_at |
AT1G19730
|
ATH4, thioredoxin H-type 4, ATTRX4 |
-0.198 |
-0.020 |
| 153 |
252685_at |
AT3G44410
|
[pseudogene] |
-0.197 |
0.028 |
| 154 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.195 |
0.162 |
| 155 |
247345_at |
AT5G63760
|
ARI15, ARIADNE 15, ATARI15, ARABIDOPSIS ARIADNE 15 |
-0.194 |
-0.048 |
| 156 |
261451_at |
AT1G21060
|
unknown |
-0.192 |
0.020 |
| 157 |
256212_at |
AT1G50970
|
[Membrane trafficking VPS53 family protein] |
-0.191 |
-0.003 |
| 158 |
262136_at |
AT1G77850
|
ARF17, auxin response factor 17 |
-0.190 |
-0.020 |
| 159 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.188 |
0.155 |
| 160 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
-0.188 |
-0.080 |
| 161 |
251057_at |
AT5G01780
|
[2-oxoglutarate-dependent dioxygenase family protein] |
-0.187 |
0.004 |
| 162 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
-0.186 |
0.007 |
| 163 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.186 |
0.005 |
| 164 |
259000_at |
AT3G01860
|
unknown |
-0.183 |
-0.003 |
| 165 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
-0.183 |
0.043 |
| 166 |
262178_at |
AT1G77860
|
KOM, KOMPEITO |
-0.181 |
-0.010 |
| 167 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.181 |
0.076 |
| 168 |
262797_at |
AT1G20840
|
AtTMT1, TMT1, tonoplast monosaccharide transporter1 |
-0.179 |
0.008 |
| 169 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.179 |
-0.006 |
| 170 |
249261_at |
AT5G41710
|
[Mutator-like transposase family, has a 6.2e-18 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.178 |
-0.017 |
| 171 |
262049_at |
AT1G80180
|
unknown |
-0.178 |
0.025 |
| 172 |
262204_at |
AT2G01100
|
unknown |
-0.178 |
0.016 |
| 173 |
256099_at |
AT1G13710
|
CYP78A5, cytochrome P450, family 78, subfamily A, polypeptide 5, KLU, KLUH |
-0.177 |
-0.042 |
| 174 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
-0.175 |
0.087 |
| 175 |
247828_at |
AT5G58510
|
unknown |
-0.174 |
0.022 |
| 176 |
247792_at |
AT5G58787
|
[RING/U-box superfamily protein] |
-0.173 |
0.117 |
| 177 |
245572_at |
AT4G14720
|
PPD2, PEAPOD 2, TIFY4B |
-0.170 |
0.022 |
| 178 |
251869_at |
AT3G54500
|
LNK2, night light-inducible and clock-regulated 2 |
-0.167 |
-0.016 |
| 179 |
246759_at |
AT5G27950
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.166 |
0.029 |
| 180 |
261398_at |
AT1G79610
|
ATNHX6, ARABIDOPSIS NA+/H+ ANTIPORTER 6, NHX6, Na+/H+ antiporter 6 |
-0.165 |
-0.047 |
| 181 |
258647_at |
AT3G07870
|
[F-box and associated interaction domains-containing protein] |
-0.163 |
0.034 |
| 182 |
250167_at |
AT5G15310
|
ATMIXTA, ATMYB16, myb domain protein 16, MYB16, myb domain protein 16 |
-0.162 |
-0.016 |
| 183 |
247890_at |
AT5G57940
|
ATCNGC5, cyclic nucleotide gated channel 5, CNGC5, cyclic nucleotide gated channel 5 |
-0.162 |
0.016 |
| 184 |
248256_at |
AT5G53360
|
[TRAF-like superfamily protein] |
-0.159 |
0.045 |
| 185 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.158 |
-0.010 |
| 186 |
245022_at |
ATCG00560
|
PSBL, photosystem II reaction center protein L |
-0.157 |
-0.003 |
| 187 |
252157_at |
AT3G50430
|
unknown |
-0.156 |
0.043 |
| 188 |
262813_at |
AT1G11670
|
[MATE efflux family protein] |
-0.156 |
0.009 |
| 189 |
261815_at |
AT1G08320
|
bZIP21, TGA9, TGACG (TGA) motif-binding protein 9 |
-0.156 |
-0.014 |
| 190 |
263898_at |
AT2G21950
|
SKIP6, SKP1 interacting partner 6 |
-0.155 |
-0.005 |
| 191 |
256058_at |
AT1G07240
|
UGT71C5, UDP-glucosyl transferase 71C5 |
-0.153 |
0.060 |
| 192 |
263542_at |
AT2G21630
|
[Sec23/Sec24 protein transport family protein] |
-0.152 |
0.024 |
| 193 |
259583_at |
AT1G28070
|
unknown |
-0.152 |
-0.003 |